Keywords
Antibiotic resistance, mecA, MRSA, PVL, Staphylococcus aureus
This article is included in the Cell & Molecular Biology gateway.
Staphylococcus aureus infections, including Methicillin-Resistant S. aureus (MRSA) and Methicillin-Sensitive S. aureus (MSSA), present significant challenges in healthcare due to rising antimicrobial resistance. This study evaluates the genetic basis of antibiotic resistance in S. aureus, focusing on key resistance-associated genes mecA and PVL.
A total of 568 clinical specimens were analyzed for the presence of S. aureus. Demographic data were collected to assess age-dependent prevalence. Antimicrobial susceptibility testing was conducted to evaluate resistance patterns. The prevalence of the mecA and PVL genes was determined using molecular techniques.
S. aureus was identified in 37.9% of cases, with the highest prevalence (60-79 age group). All S. aureus isolates showed 100% resistance to penicillin. Multidrug-resistant (MDR) strains accounted for 84.19% of isolates, with a significant presence of extensively drug-resistant (XDR) strains. The mecA gene was prevalent in 82.79% of MRSA isolates, indicating its strong association with methicillin resistance. Additionally, 41.86% of all S. aureus isolates were positive for the PVL gene, highlighting its widespread distribution.
The high prevalence of mecA and PVL genes in S. aureus strains underscores the challenges in managing these infections. These findings emphasize the necessity for judicious antibiotic use and enhanced collaborative efforts to combat antimicrobial resistance. Understanding the genetic basis of resistance can inform more effective diagnostic, therapeutic, and preventive strategies, ultimately improving patient outcomes in S. aureus infections.
Antibiotic resistance, mecA, MRSA, PVL, Staphylococcus aureus
Staphylococcus aureus presents in two major variants, Methicillin-Resistant S. aureus (MRSA) and Methicillin-Sensitive S. aureus (MSSA), each posing distinct challenges and requiring tailored approaches for management. MRSA, resilient against methicillin and diverse beta-lactam antibiotics, poses a significant global health threat, leading to severe complications and increased mortality within 48 hours of hospital admission (Chatterjee et al., 2018; Rubio-Garcia et al., 2023). Innovative treatment strategies, such as combining natural compounds with antibiotics, have been explored to enhance bactericidal efficacy and minimize side effects, focusing on preventing MRSA introduction and dissemination in healthcare and community settings (Al-Tawalbeh et al., 2023; Woźniak et al., 2024). Effective MRSA prevention necessitates the identification and control of risk factors, including prolonged surgery duration, extensive antibiotic use, and patient condition (Sun et al., 2024a).
Conversely, MSSA, susceptible to methicillin and other beta-lactam antibiotics, coexists with MRSA, demanding attention to different therapeutic considerations (Arif et al., 2024). While challenges arise from increasing antimicrobial resistance in MSSA, the primary focus remains on MRSA mitigation. Conventional beta-lactam agents, particularly cefazolin, have demonstrated efficacy, with considerations for central nervous system infections and reduced administration barriers compared to penicillins (Antosz et al., 2023). Persistent MSSA bacteremia complexities require source control and modified therapy, with combination therapy and recognition of the inoculum effect showing promise (Chastain et al., 2023). Strategies for MSSA prevention involve addressing resistance challenges in decolonization, utilizing agents like mupirocin, and employing rapid diagnostic tests for tailored treatment (Palavecino, 2020). The development of a protective vaccine holds potential to reduce morbidity and mortality associated with invasive MSSA disease, forming a comprehensive strategy against these pathogens (Cassat & Thomsen, 2021).
The research aims to evaluate the current status of antibiotic resistance in S. aureus, focusing on the key resistance-associated genes, mecA and PVL. The mecA gene, critical in methicillin resistance, plays a central role in the evolution of MRSA, rendering it resilient against beta-lactam antibiotics (John, 2020; Tasneem et al., 2022). Understanding the genetic organization of mecA, as emphasized by Abdullahi et al. (2023), is crucial in addressing multidrug resistance challenges, particularly in non-aureus staphylococci.
PVL stands out as a virulence factor in S. aureus, significantly contributing to pathogenicity and rapid dissemination, especially in skin and soft tissue infections (Leistner et al., 2022; Tromp & van Strijp, 2020). The research will explore the role of PVL in the increased virulence and epidemic spread of S. aureus, aiming to unravel the exact mechanisms and contributions to the pathophysiology of this versatile bacterium. Diagnostic and therapeutic aspects related to PVL will be investigated, considering its association with recurrent skin infections and potential links to broader health issues, including cancer (Wei et al., 2022). The study endeavors to contribute to a comprehensive understanding of the impact of these key genes on S. aureus resistance and pathogenicity.
MRSA and MSSA pose significant challenges in the control and prevention of S. aureus infections. While MRSA presents grave threats due to extensive antibiotic resistance, MSSA shows rising drug resistance. Key resistance and virulence genes such as mecA and PVL play pivotal roles in the evolution and pathogenicity of S. aureus. Further investigation into these genetic mechanisms will support diagnostic, therapeutic, and preventative strategies to effectively control MRSA.
S. aureus strains were isolated from clinical samples obtained from patients at Can Tho University of Medicine and Can Tho City General Hospital. The sampling process involved patients with S. aureus infections, and the bacterial isolates were identified using the Vitek system, following the methodology outlined by Tran et al. (2023).
Various types of clinical specimens were collected from different departments, including the Internal Medicine Department (IMD), Trauma Surgery Department (TSD), Surgical Department (SD), Outpatient Clinic Department (OCD), as well as other specialized departments such as Oncology Department (OD), Cardiology Department (CD), Nephrology and Urology Department (NUD), and External Medicine Department (EMD). The collected specimens encompassed pus, phlegm, fluid, and blood, providing a comprehensive representation of bacterial strains in diverse clinical conditions.
The sampling duration extended from April 2022 to April 2023, ensuring a year-long representation of S. aureus strains prevalent in the patient population during this period.
Antimicrobial susceptibility testing of S. aureus isolates was facilitated utilizing the Vitek-2 Compact automated system (bioMérieux, USA). The AST-GN67 testing card (Catalog number: 413399; bioMérieux, USA) enabled efficient determination of minimum inhibitory concentrations for selected antibiotics based on CLSI breakpoints. CLSI, antibiotic susceptibility interpretation covered agents across categories, with details provided in Table 1. Isolates were classified by resistance patterns - multidrug-resistant (MDR), extensively drug-resistant (XDR) or pan-drug-resistant (PDR), applying standard definitions of non-susceptibility to one/three or more agents (Tran et al., 2023). This approach leveraging automated technology ensured rigorous evaluation of antibiotic resistance across S. aureus isolates within a streamlined workflow.
The mecA, and PVL genes were detected in all S. aureus isolates. Genomic DNA extraction from the cells was performed using the Invisorb® Spin Universal kit (Catalog number: 1050100300; Invitek Molecular, Germany) (Tran et al., 2023). PCR amplification was conducted using the MyTaq™ Red Mix Kit (Catalog number: BIO-25044; Meridian Bioscience, USA) and the C1000 Touch Thermal Cycler system (Bio-Rad, USA). The reaction mixture included specific primers for each gene, as detailed in Table 2 (Hsu et al., 2004; Le et al., 2021; Lee, 2003). The PCR amplification conditions comprised an initial denaturation step at 95°C for 3 minutes, followed by 40 cycles of amplification at 95°C for 30 seconds, 55°C for 30 seconds, and 72°C for 30 seconds. The final extension step was maintained at 72°C for 5 minutes.
T-tests were used to compare mean differences between two groups, such as different testing methods. Analysis of Variance (ANOVA) was employed to assess differences among three or more groups, such as various testing methods. Additionally, multivariate analysis was utilized to evaluate relationships between multiple variables simultaneously. These methods ensured thorough analysis of the data and provided insights into the efficacy of different testing methods and correlations between variables.
The study involved the analysis of 568 clinical specimens, with the Vitek-2 Compact automated system used for automatic identification. Results from this identification process revealed the presence of 215 out of 568 isolates as S. aureus, constituting 37.9% of the total samples (p < 0.05). Additionally, E. coli and K. pneumoniae were identified at rates of 25.7% and 21.2% (p = 0.05).
To provide a demographic perspective, the age distribution of individuals with S. aureus infections is illustrated in Figure 1. The age range for male patients varied from 28 to 101 years, with a median age of 68 and a mean age of 67.78. On the other hand, female patients exhibited an age range from 20 to 102 years, with a median age of 67 and a mean age of 66.76. These findings offer insights into the age and gender distribution of individuals affected by S. aureus infections in the studied population (p > 0.05).
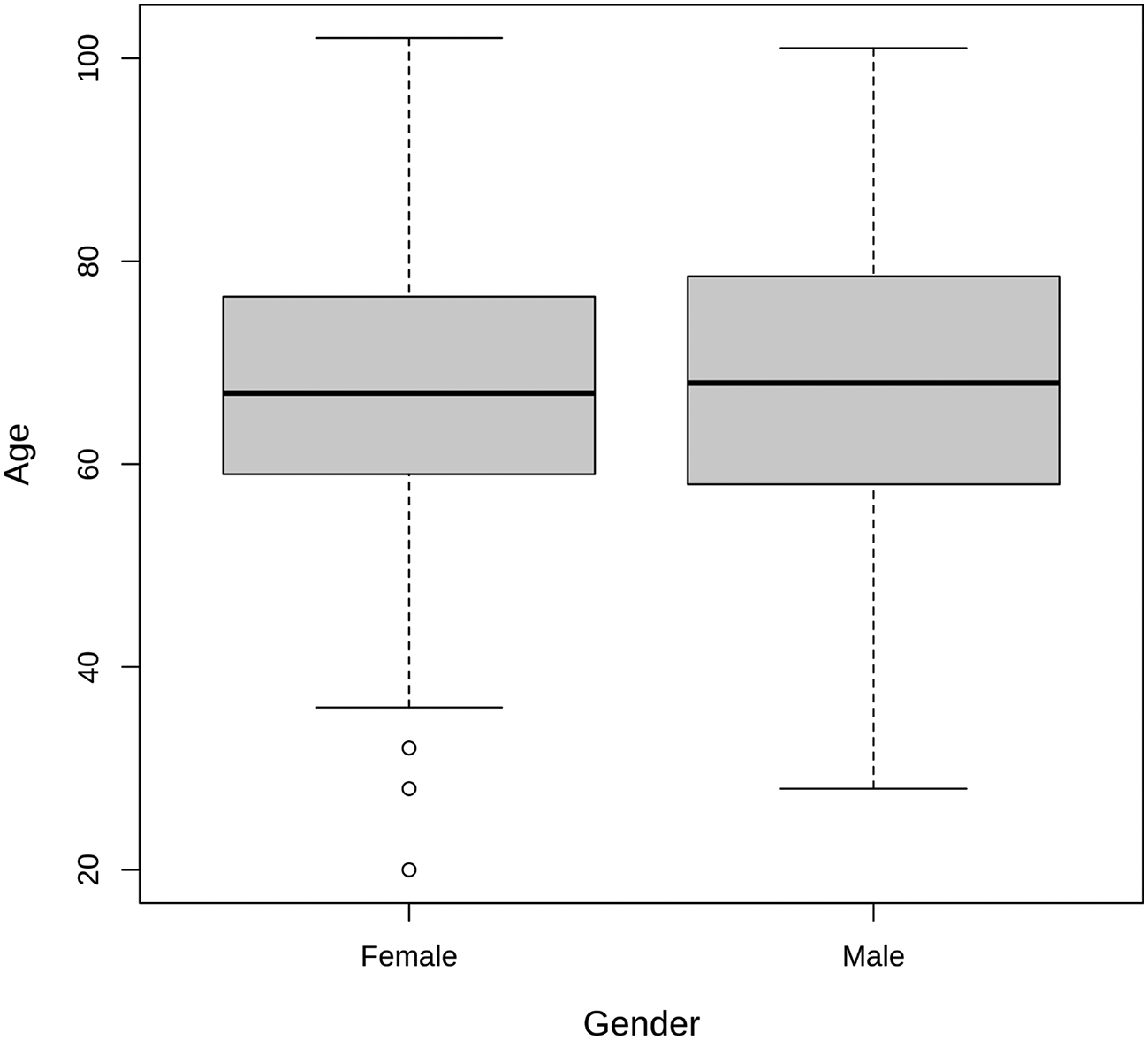
Atl text: The age range for male patients varied from 28 to 101 years, with a median age of 68 and a mean age of 67.78. For female patients, the age range was from 20 to 102 years, with a median age of 67 and a mean age of 66.76. This figure provides insights into the age and gender distribution of individuals affected by S. aureus infections in the studied population (p > 0.05).
For the age group 20-39, the presence of S. aureus was observed in 5 males (2.33%) and 6 females (2.79%). In the 40-59 age group, the prevalence increased, with 27 males (12.56%) and 21 females (9.77%) testing positive for S. aureus (Figure 2). The age group 60-79 exhibited the highest prevalence, with 66 males (30.70%) and 42 females (19.53%) found to have S. aureus infections. Among individuals over 80 years old, 26 males (12.09%) and 22 females (10.23%) were identified with S. aureus (Figure 2). These results highlight an age-dependent pattern, with a notable increase in S. aureus prevalence in the older age groups, particularly in individuals aged 60-79. This information contributes valuable insights into the distribution of S. aureus infections across different age categories (p < 0.05), aiding in the understanding of demographic factors associated with susceptibility to these infections.
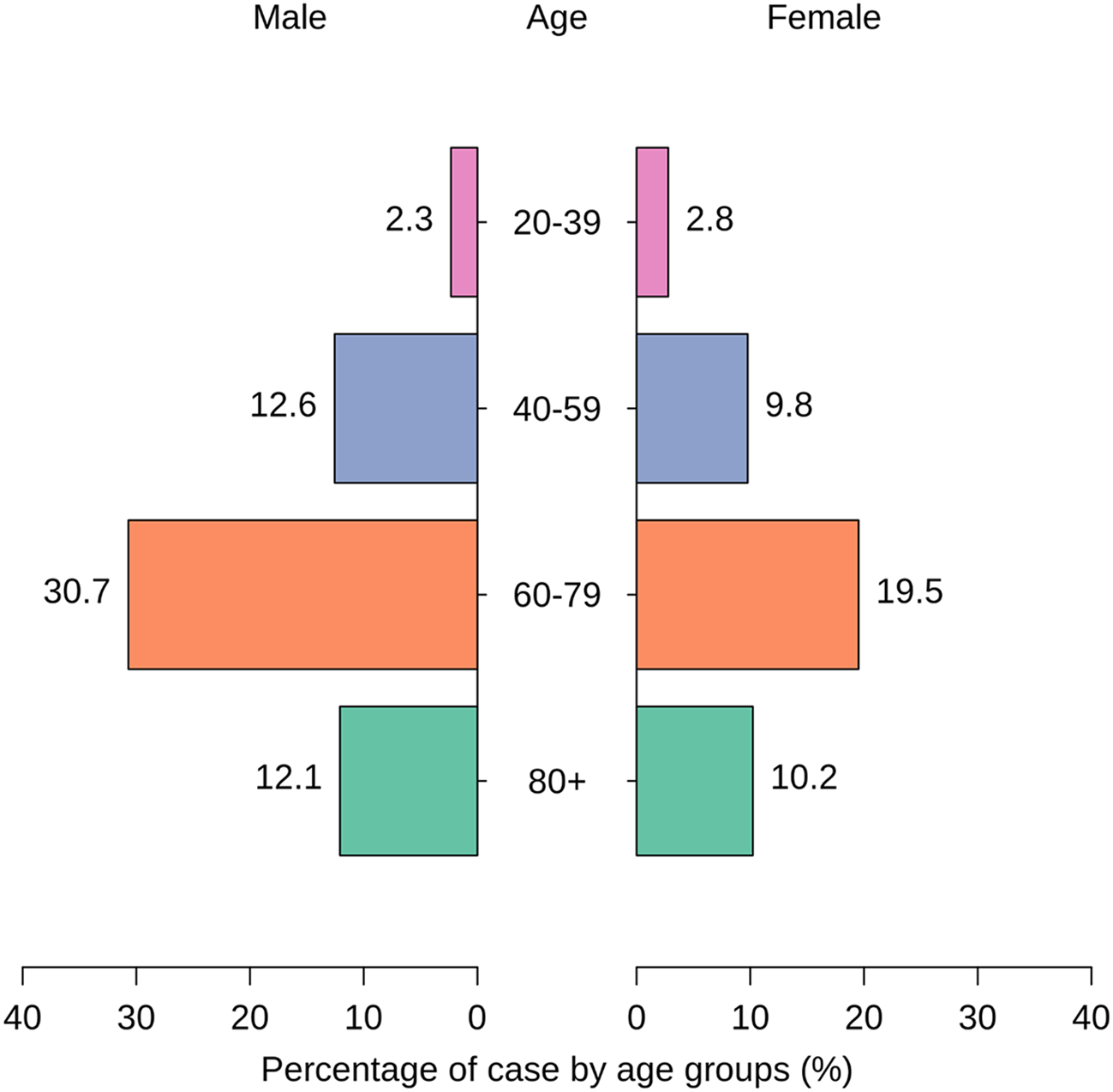
Atl text: In the 20-39 age group, 5 males (2.33%) and 6 females (2.79%) were infected. In the 40-59 age group, the prevalence increased to 27 males (12.56%) and 21 females (9.77%). The 60-79 age group exhibited the highest prevalence with 66 males (30.70%) and 42 females (19.53%) infected. Among individuals over 80 years old, 26 males (12.09%) and 22 females (10.23%) were identified with S. aureus. These results show an age-dependent pattern, with a significant increase in prevalence in older age groups, especially those aged 60-79 (p < 0.05).
Figure 3 illustrates the distribution of S. aureus detection across different illnesses, providing valuable insights into the prevalence of S. aureus in association with specific health conditions. The majority of cases, constituting 36.28%, showed no reported illness. This suggests that a considerable proportion of individuals without underlying health issues also experienced S. aureus infections. In contrast, diabetes mellitus emerged as a significant contributing factor, with 52.56% of cases associated with this condition. The prevalence of S. aureus in individuals with renal failure accounted for 6.05%, highlighting the relevance of kidney-related health concerns. Specific medical interventions, such as open surgery, were linked to 2.79% of S. aureus cases, emphasizing the potential risk within healthcare settings. Additionally, cancer was associated with 2.33% of cases, indicating the presence of S. aureus in individuals undergoing cancer-related treatments. This detailed breakdown of illness categories provides a nuanced understanding of the varied contexts in which S. aureus infections occur. The higher prevalence in diabetes mellitus cases underscores the importance of tailored strategies for infection prevention and management in individuals with specific underlying health conditions.
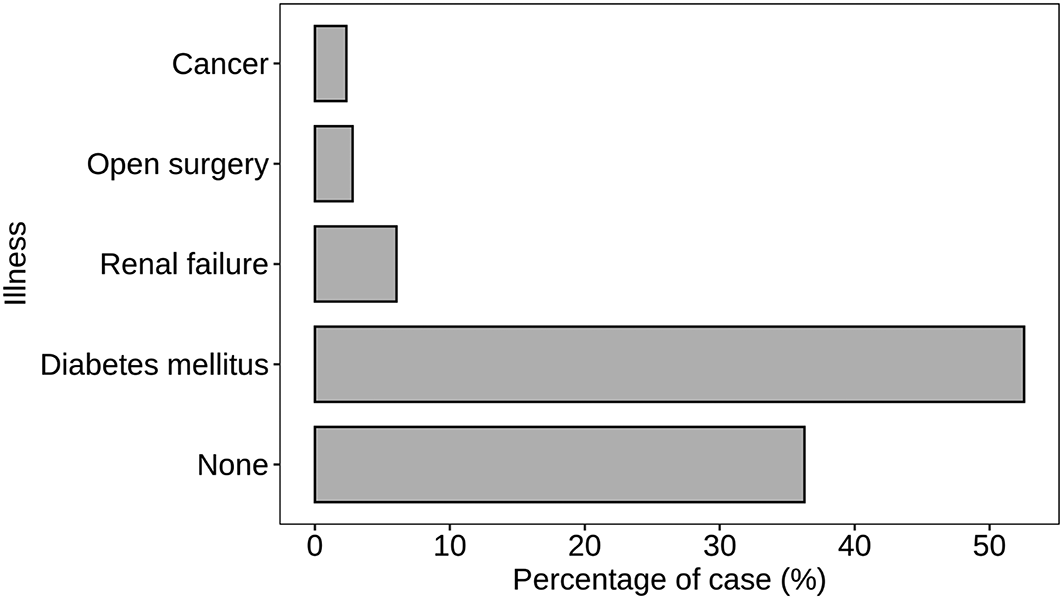
Atl text: A substantial proportion of cases (36.28%) had no reported illness, while diabetes mellitus was associated with 52.56% of cases. Renal failure accounted for 6.05% of cases, open surgery for 2.79%, and cancer for 2.33%. This figure highlights the prevalence of S. aureus in association with specific health conditions, emphasizing the need for tailored infection prevention and management strategies in individuals with certain underlying conditions.
Figure 4 presents an insightful analysis of the distribution of S. aureus detection concerning underlying health conditions. The data highlights the significance of the immunocompromised status in relation to S. aureus infections. Among the cases analyzed, 63.26% were associated with individuals classified as immunocompromised. This substantial proportion emphasizes the heightened vulnerability of individuals with compromised immune systems to S. aureus infections. It underscores the critical role of immune function in protecting against such bacterial infections. Conversely, 36.74% of cases were attributed to individuals categorized as non-immunocompromised. While this group represents a significant portion, the data underscores that S. aureus infections are not exclusive to immunocompromised individuals, as they also affect those with apparently normal immune function. The distinct division between immunocompromised and non-immunocompromised categories provides a clear understanding of the association between immune status and susceptibility to S. aureus infections (p > 0.05). This insight is crucial for tailoring preventive measures and treatment strategies based on the specific health conditions of individuals.
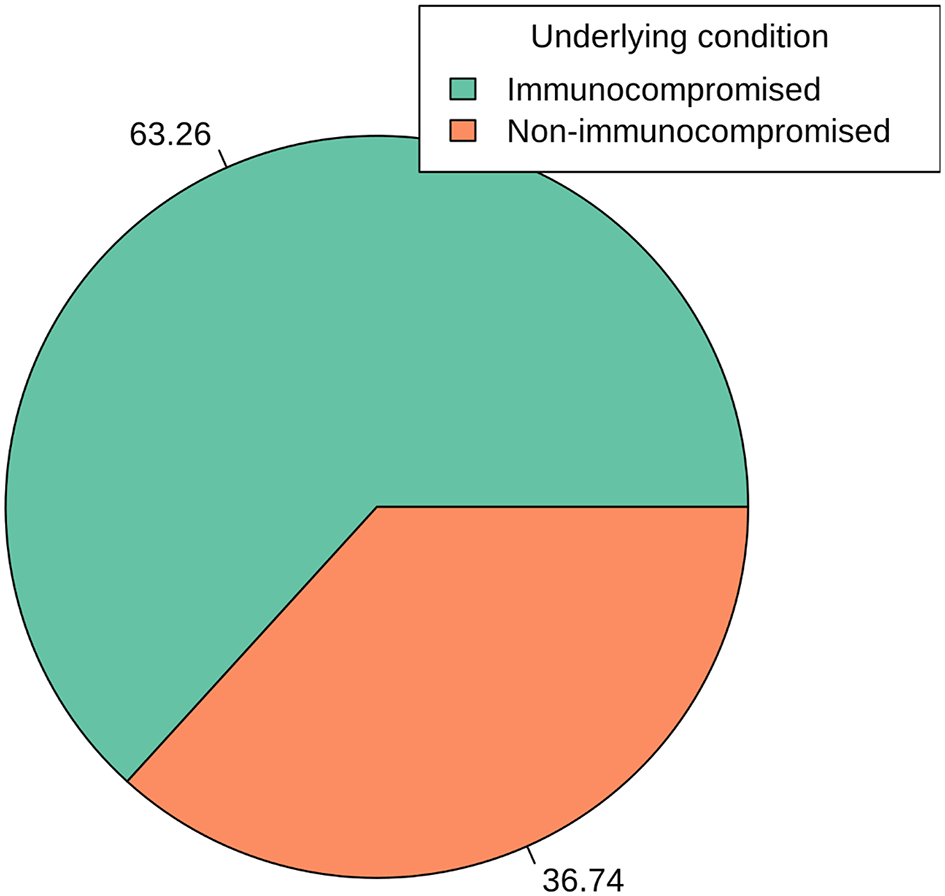
Atl text: Among the analyzed cases, 63.26% were associated with immunocompromised individuals, indicating a heightened vulnerability to S. aureus infections. Conversely, 36.74% of cases were found in non-immunocompromised individuals. This figure emphasizes the significant role of immune function in susceptibility to S. aureus infections and the need for specific preventive measures for immunocompromised individuals (p > 0.05).
Figure 5 provides a detailed breakdown of S. aureus detection across different departments within the hospital setting. The data sheds light on the prevalence of S. aureus in various medical departments, indicating potential areas of focus for infection control and preventive strategies. The Internal Medicine Department (IMD) exhibits the highest count of S. aureus cases, representing 39.53% of the total cases. This suggests a significant presence of S. aureus infections among patients seeking treatment in the Internal Medicine Department. The Trauma Surgery Department (TSD) follows closely with 20.93% of cases, emphasizing the relevance of S. aureus infections in patients undergoing trauma-related medical interventions. Surgical Department (SD) and Outpatient Clinic Department (OCD) contribute 16.28% and 15.81% to the total cases (p < 0.05), indicating a substantial presence of S. aureus in these departments. Additionally, there are 7.44% of cases categorized under “Other” departments, which may include specialty units such as Oncology, Cardiology, Nephrology and Urology, and External Medicine. This suggests a distributed occurrence of S. aureus infections in various specialized areas of the hospital. The department-wise analysis presented in Figure 5 enables healthcare professionals to target specific areas for heightened surveillance, infection prevention, and control measures tailored to the distinct characteristics of each medical department.
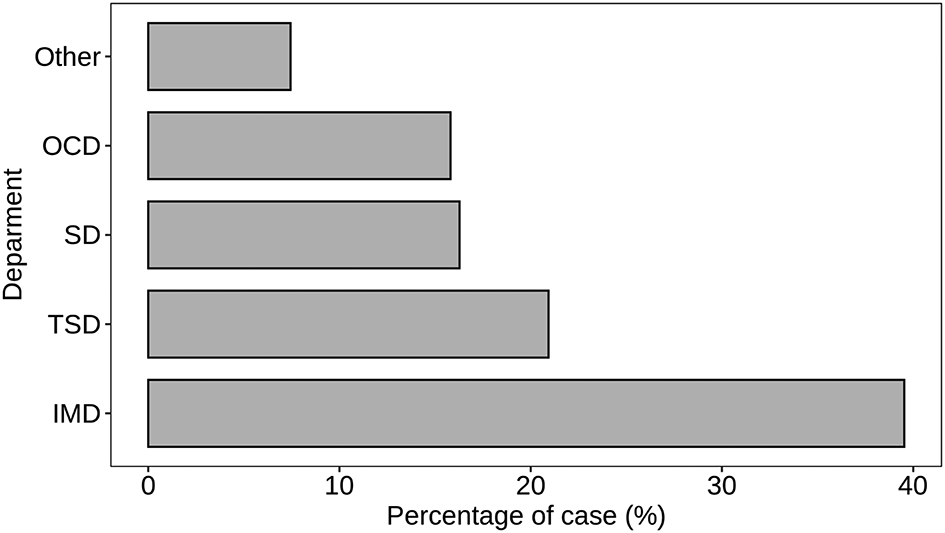
Atl text: The Internal Medicine Department (IMD) exhibited the highest count of S. aureus cases (39.53%), followed by the Trauma Surgery Department (TSD) with 20.93%. The Surgical Department (SD) and Outpatient Clinic Department (OCD) accounted for 16.28% and 15.81% of cases, respectively (p < 0.05). An additional 7.44% of cases were categorized under "Other" departments, which include specialized units such as Oncology, Cardiology, Nephrology and Urology, and External Medicine. This department-wise analysis helps target specific areas for enhanced surveillance and infection control measures.
Figure 6 illustrates the antimicrobial susceptibility patterns of S. aureus isolates across different antibiotic groups. The data provides insights into the prevalence of resistance, intermediate susceptibility, and susceptibility to various antibiotics commonly used in clinical settings.
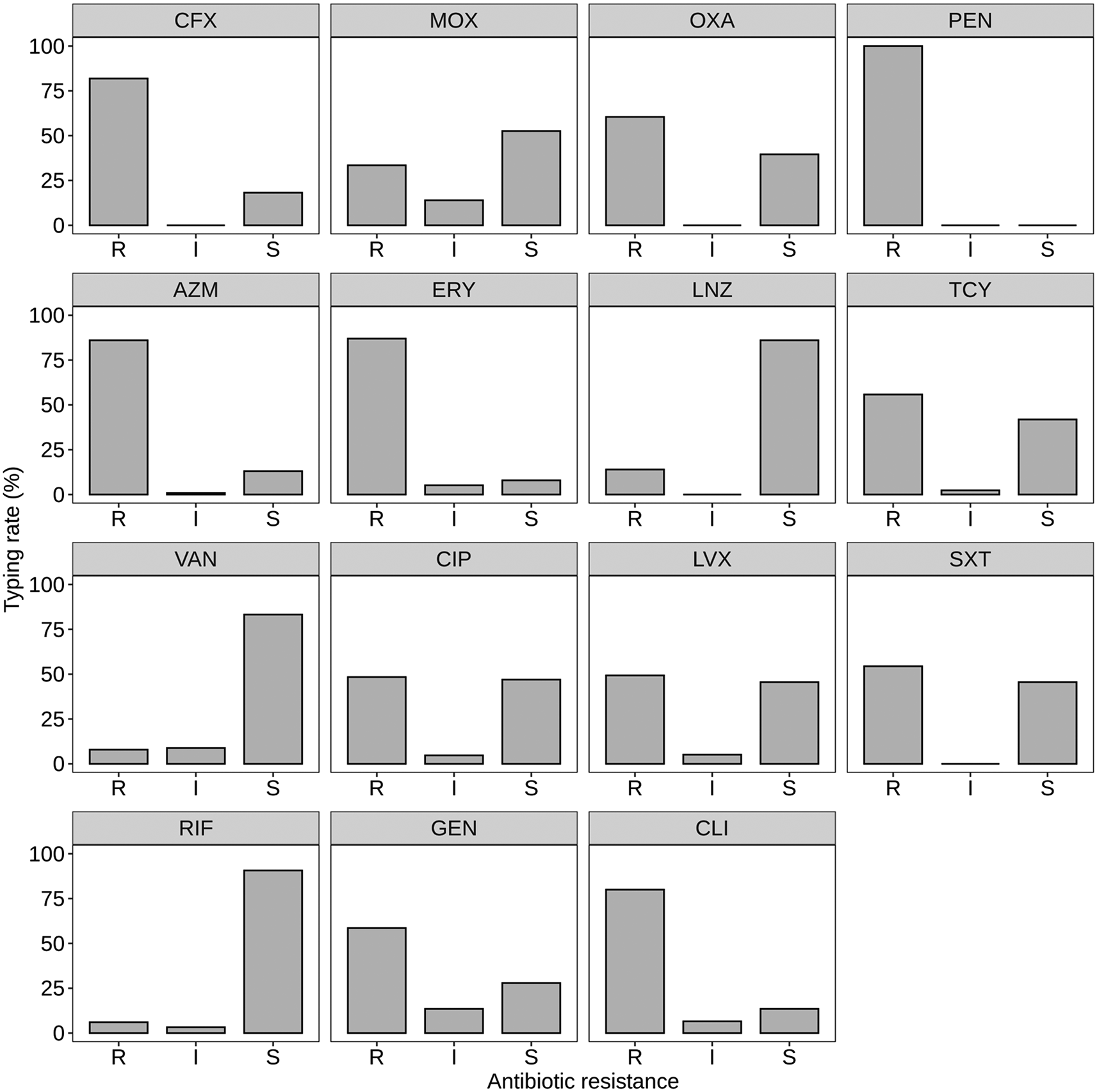
Note: R = Resistance, I = Intermediate susceptibility, S = Susceptibility. Atl text: The majority of S. aureus isolates exhibited 100% resistance to penicillin (PEN). Resistance rates varied for other beta-lactam antibiotics: cefoxitin (CFX) showed 81.86% resistance, oxacillin (OXA) 60.46%, and amoxicillin-clavulanate (MOX) 33.48% (p < 0.05). No intermediate susceptibility was observed for beta-lactam antibiotics. For macrolides, high resistance was noted: azithromycin (AZM) at 86.04% and erythromycin (ERY) at 86.97% (p < 0.05). Among peptid antibiotics, linezolid (LNZ) had the highest susceptibility at 86.05%, while tetracycline (TCY) and vancomycin (VAN) showed resistance rates of 55.81% and 7.91%, respectively (p < 0.05). In the quinolone group, ciprofloxacin (CIP) and levofloxacin (LVX) had resistance rates of 48.37% and 49.3%, with sulfamethoxazole-trimethoprim (SXT) showing 54.41% resistance (p < 0.05). Other antibiotics showed varying resistance rates: rifampicin (RIF) at 6.05%, gentamicin (GEN) at 58.6%, and clindamycin (CLI) at 80% (p < 0.05). The data underscores widespread resistance to multiple antibiotic classes, emphasizing the need for careful antibiotic use and ongoing resistance monitoring.
For beta-lactam antibiotics, the majority of S. aureus isolates exhibited resistance to penicillin (PEN), with 100% resistance observed. Resistance rates to other beta-lactam antibiotics varied, with cefoxitin (CFX) showing 81.86% resistance, oxacillin (OXA) 60.46% resistance, and amoxicillin-clavulanate (MOX) 33.48% resistance (p < 0.05). Intermediate susceptibility was not observed for any of the beta-lactam antibiotics tested.
In the macrolide group, S. aureus isolates showed high resistance to azithromycin (AZM) at 86.04% and erythromycin (ERY) at 86.97%. Intermediate susceptibility was minimal, while susceptibility rates were relatively low for both antibiotics (p < 0.05).
Among the peptid antibiotics, linezolid (LNZ) demonstrated the highest susceptibility, with 86.05% of isolates being susceptible. Resistance rates were notable for tetracycline (TCY) at 55.81% and vancomycin (VAN) at 7.91%, with intermediate susceptibility observed for both antibiotics (p < 0.05).
In the quinolone group, ciprofloxacin (CIP) and levofloxacin (LVX) exhibited comparable resistance rates, with 48.37% and 49.3% resistance. Sulfamethoxazole-trimethoprim (SXT) showed resistance in 54.41% of isolates. Intermediate susceptibility was observed for some isolates across these antibiotics (p < 0.05).
For other antibiotics, rifampicin (RIF) displayed resistance in 6.05% of isolates, while gentamicin (GEN) and clindamycin (CLI) exhibited resistance rates of 58.6% and 80%, respectively. Intermediate susceptibility was noted for rifampicin, gentamicin, and clindamycin, with varying proportions (p < 0.05).
Overall, the data underscores the widespread resistance of S. aureus isolates to multiple classes of antibiotics, emphasizing the importance of judicious antibiotic use and surveillance to combat antimicrobial resistance.
Figure 7 depicts the distribution of resistance patterns among S. aureus isolates. The data reveals three main resistance patterns: multidrug-resistant (MDR), extensively drug-resistant (XDR), and non-resistant. Multidrug-resistant (MDR) strains constitute the majority, with 181 isolates, accounting for 84.19% of the total. These strains exhibit resistance to antibiotics from multiple classes, posing significant challenges in treatment selection. Extensively drug-resistant (XDR) strains, characterized by resistance to most available antibiotics except a few, are observed in 10 isolates, representing 4.65% of the total. XDR strains present even greater therapeutic challenges due to limited effective treatment options. Non-resistant strains, comprising 24 isolates (11.16%), are susceptible to the antibiotics tested in this study (p < 0.05). While these strains are susceptible to antimicrobial agents, monitoring their susceptibility patterns remains crucial to prevent the emergence of resistance.
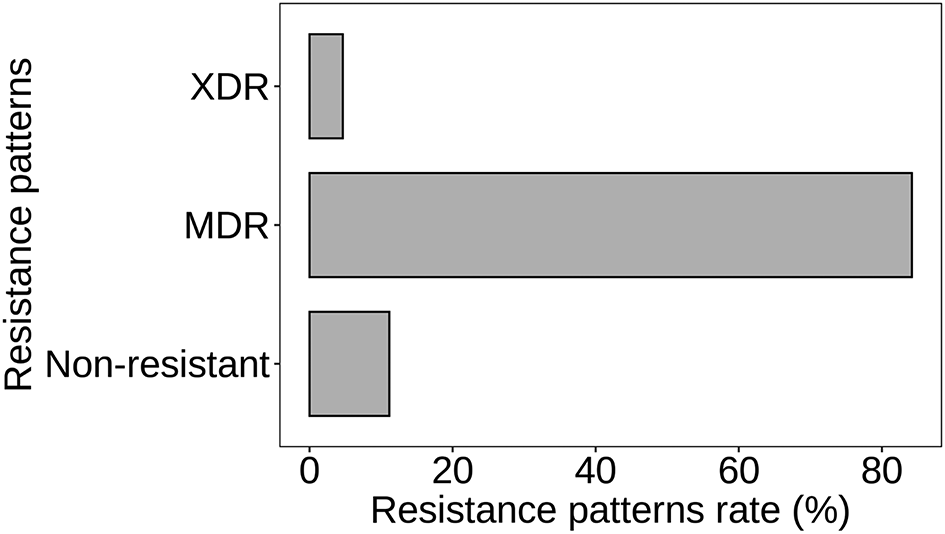
Atl text: The data reveals three main resistance patterns: multidrug-resistant (MDR), extensively drug-resistant (XDR), and non-resistant. MDR strains constituted the majority with 181 isolates (84.19%), showing resistance to multiple antibiotic classes. XDR strains, resistant to most antibiotics except a few, accounted for 10 isolates (4.65%), presenting significant treatment challenges. Non-resistant strains, comprising 24 isolates (11.16%), were susceptible to the antibiotics tested in this study (p < 0.05). Monitoring susceptibility patterns of these non-resistant strains remains crucial to prevent the development of resistance.
In Figure 8A, a detailed analysis of mecA gene detection among S. aureus isolates is presented, including both count and proportion of detections. Of the total isolates analyzed, 178 (82.79%) tested positive for the mecA gene, indicating the presence of methicillin resistance mechanisms. Conversely, 37 isolates (17.21%) did not exhibit mecA gene detection, suggesting the absence of this specific genetic determinant associated with methicillin resistance. Further analysis based on methicillin resistance status reveals that mecA gene detection was prevalent among MRSA isolates, with 67.91% testing positive for the gene. This underscores the strong association between mecA gene presence and methicillin resistance. Among MRSA-negative cases, mecA gene detection was considerably lower, with only 14.88% of isolates testing positive. Interestingly, a similar proportion of MRSA-negative cases also exhibited mecA gene detection, suggesting potential discrepancies or alternative mechanisms of methicillin resistance in these isolates. Additionally, the absence of mecA gene detection was observed in 2.33% of MRSA-negative cases, indicating a small subset of isolates without mecA-associated methicillin resistance.
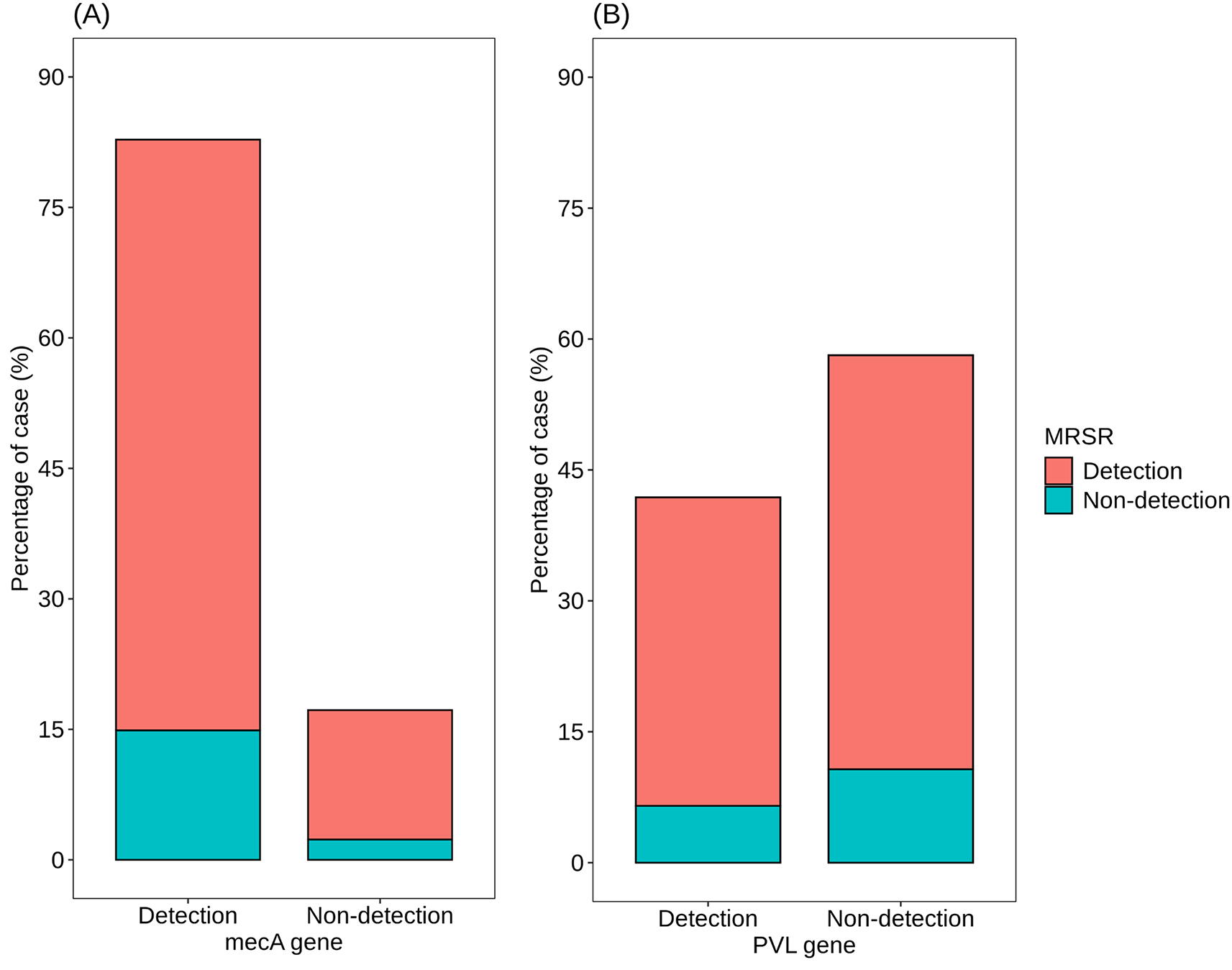
Atl text: (A) Detailed analysis of mecA gene detection. Among the total isolates analyzed, 178 (82.79%) tested positive for the mecA gene, indicating methicillin resistance mechanisms. Conversely, 37 isolates (17.21%) were negative for the mecA gene. Analysis based on methicillin resistance status reveals that 67.91% of MRSA isolates tested positive for the mecA gene, underscoring the association between mecA presence and methicillin resistance. Among MRSA-negative cases, 14.88% were positive for the mecA gene, suggesting potential alternative resistance mechanisms. Additionally, 2.33% of MRSA-negative cases showed no mecA detection, indicating the absence of this methicillin resistance determinant in a small subset of isolates. (B) Comprehensive analysis of PVL gene detection. Among all isolates analyzed, 90 (41.86%) were positive for the PVL gene, indicating its prevalence among the sampled S. aureus strains. Conversely, 125 isolates (58.14%) were negative for the PVL gene. Examination based on methicillin resistance status shows that 35.35% of MRSA isolates had the PVL gene, while 6.51% of MRSA-negative cases were positive for the gene. Interestingly, 47.44% of non-MRSA isolates exhibited the presence of the PVL gene, indicating its occurrence in a significant proportion of methicillin-sensitive S. aureus strains. Additionally, 10.7% of non-MRSA isolates did not detect the PVL gene.
In Figure 8B, a comprehensive analysis of PVL gene detection among S. aureus isolates is provided, considering both count and proportion of detections. Among all isolates analyzed, 90 (41.86%) tested positive for the presence of the PVL gene, indicating its prevalence among the sampled S. aureus strains. Conversely, 125 isolates (58.14%) did not exhibit detection of the PVL gene. Further examination based on methicillin resistance status shows that among MRSA isolates, the detection of the PVL gene was observed in 35.35% of cases, indicating its presence in a significant proportion of methicillin-resistant strains. In contrast, among MRSA-negative cases, the detection rate of the PVL gene was notably lower, with only 6.51% of isolates testing positive. This suggests a reduced prevalence of the PVL gene among methicillin-sensitive S. aureus strains. Interestingly, a considerable percentage of non-MRSA isolates exhibited the presence of the PVL gene, with 47.44% of MRSA-negative cases testing positive for its detection. This indicates that the PVL gene is not exclusive to MRSA strains and is also present in a substantial proportion of methicillin-sensitive S. aureus strains. Additionally, 10.7% of non-MRSA isolates did not show detection of the PVL gene.
Al-Tawalbeh et al. (2023) proposed a novel approach in MRSA treatment by combining carvacrol - a component of essential oils, with approved antibiotics. Carvacrol has demonstrated antimicrobial activity against both gram-positive and gram-negative bacteria. The study revealed that the combination of carvacrol with antibiotics provided better efficacy compared to monotherapy, particularly showing superior results with linezolid, minocycline, and sulfamethoxazole. This suggests potential synergistic or additive effects between carvacrol and certain antibiotics. Specifically, among MRSA isolates, carvacrol combined with linezolid, minocycline, and sulfamethoxazole exhibited the following proportions of detections: 87.3%, 89.5%, and 84.2%. In the study by Rubio-Garcia et al. (2023), some antiretroviral drugs exhibited antibacterial activity against human commensal bacteria and multidrug-resistant pathogens, including MRSA. This highlights the potential repurposing of antiretroviral drugs for treating bacterial infections, particularly in cases of multidrug resistance. Notably, elvitegravir showed antibacterial activity against G. vaginalis and P. bivia, as well as against vancomycin-resistant Enterococcus spp. and MRSA strains, with MIC values ranging from 4 to 16 μg/mL. Sun et al. (2024b) emphasized the role of antibiotic misuse in the rise of multidrug-resistant organisms (MDROs) and identified factors contributing to postoperative MDRO infections. Their findings underscored the importance of judicious antibiotic use, shorter operative times, and increased patient mobility in preventing surgical-site infections with MDROs. Furthermore, among patients with limb fractures, the risk factors for MDRO infection included being bedridden (OR, 2.66; P = 0.037), repeated infection (OR, 4.00; P = 0.005), and an operative time of >3 h (OR, 2.28; P = 0.023). Kungu et al. (2024) revealed the prevalence of MRSA among humans and animals in Uganda. Their study highlighted the need for surveillance of antimicrobial resistance and promotion of rational antibiotic use to mitigate the spread of resistant strains. Among human and cattle samples, S. aureus isolates showed high levels of resistance to Nitroimidazoles (100%) and Penicillins (93.3%). Additionally, 93% of human isolates exhibited multidrug resistance (MDR). Islam (2024) evaluated the efficacy of vancomycin AUC-based dosing for non-MRSA infections. While AUC-based dosing may be necessary for MRSA cases, its impact on non-MRSA infections was less pronounced. In patients with non-MRSA infections, the overall early response rate to vancomycin AUC-based dosing was 50.3%, with a 30-day all-cause mortality rate of 11.3% and a rate of acute kidney injury (AKI) of 3.8%. Woźniak et al. (2024) demonstrated that eliminating environmental reservoirs and implementing improved infection control measures in the ICU reduced the incidence of infections caused by MDROs. This study emphasizes the importance of comprehensive infection control strategies in healthcare settings. Infections with MDR strains, particularly Klebsiella pneumoniae NDM, were observed to spread from hand-wash basins in wards and from inside air conditioners on the ceiling outside patient rooms.
The study conducted provides a comprehensive analysis of both antibiotic resistance patterns and demographic factors associated with S. aureus infections. By examining 568 clinical specimens, the study revealed a substantial prevalence of S. aureus, constituting 37.9% of the total samples. This high prevalence underscores the significance of understanding both the microbial and host-related determinants of S. aureus infections. Demographic analysis revealed an age-dependent pattern, with a notable increase in S. aureus prevalence among older age groups, particularly individuals aged 60-79. These findings suggest that age plays a crucial role in susceptibility to S. aureus infections, highlighting the importance of age-specific preventive strategies and targeted surveillance efforts. Furthermore, the study delved into the association between S. aureus infections and underlying health conditions, with diabetes mellitus emerging as a significant contributing factor. Over half of the cases were associated with diabetes mellitus, emphasizing the need for tailored infection prevention measures for individuals with specific comorbidities. Immunocompromised status was identified as another key determinant of S. aureus infections, with a substantial proportion of cases (63.26%) occurring in immunocompromised individuals. This underscores the heightened vulnerability of immunocompromised patients to S. aureus infections and emphasizes the critical role of immune function in protecting against bacterial pathogens. In addition to demographic factors, the study investigated the distribution of S. aureus infections across different hospital departments. The Internal Medicine Department (IMD) exhibited the highest count of S. aureus cases, suggesting a significant presence of S. aureus infections among patients seeking treatment in this department. This highlights the importance of targeted infection control measures in high-prevalence medical departments. Turning to antibiotic resistance patterns, the study revealed widespread resistance of S. aureus isolates to multiple classes of antibiotics, particularly beta-lactams and macrolides. The predominance of multidrug-etresistant (MDR) strains, comprising 84.19% of cases, underscores the challenges in treatment selection and the urgent need for antimicrobial stewardship programs. Moreover, the emergence of extensively drug-resistant (XDR) strains further complicates treatment options, emphasizing the pressing need for novel therapeutic approaches and surveillance strategies to combat antimicrobial resistance.
In conclusion, the integration of antibiotic resistance patterns and demographic factors provides a holistic understanding of S. aureus infections, enabling the development of targeted interventions to mitigate the spread of antimicrobial resistance and improve clinical outcomes. By addressing both microbial and host-related determinants, healthcare professionals can optimize infection control measures and treatment strategies to effectively manage S. aureus infections in diverse patient populations.
S. aureus, particularly MRSA, poses a significant challenge in healthcare settings due to its ability to resist multiple antibiotics. The emergence of antibiotic resistance, especially to beta-lactam antibiotics, has complicated treatment strategies over the years (John, 2020). The mecA gene, carried within the SCCmec cassette, is a key determinant of methicillin resistance in S. aureus. It encodes penicillin-binding protein 2a (PBP2a), which alters the peptidoglycan synthesis pathway, rendering the bacteria resistant to beta-lactam antibiotics (Tasneem et al., 2022). This genetic mechanism allows MRSA strains to proliferate even in the presence of beta-lactam antibiotics, leading to prolonged hospital stays and increased mortality rates (Algammal et al., 2020). Studies have highlighted the clinical manifestations and epidemiology of MRSA infections, emphasizing the need for novel treatment strategies (Shoaib et al., 2022). While vancomycin remains a standard treatment option, the rise of resistance underscores the urgency for alternative therapies (John, 2020). Several newer agents, including trimethoprim-sulfamethoxazole, ceftaroline, daptomycin, and linezolid, have shown promising activity against resistant staphylococci (John, 2020). Understanding the molecular determinants of antibiotic resistance in MRSA, particularly the role of mecA-encoded PBP2a, is crucial for developing effective treatment approaches (Lade & Kim, 2023). Furthermore, surveillance of mecA-mediated resistance in non-aureus staphylococci and mammaliicocci highlights the importance of One Health approaches in combating antibiotic resistance (Abdullahi et al., 2023). The spread of mecA/mecC-carrying strains, particularly in mastitis cases, underscores the need for vigilant monitoring and control measures in veterinary settings (Abdullahi et al., 2023). While the mecA gene plays a central role in conferring methicillin resistance, other genetic determinants and virulence factors contribute to the complexity of MRSA infections (Akoua-Koffi et al., 2022). Understanding the interplay between these factors and their impact on antibiotic resistance is essential for devising comprehensive treatment and prevention strategies. In conclusion, the mecA gene significantly influences the antibiotic resistance profile of S. aureus, particularly MRSA. Further research into the molecular mechanisms underlying resistance and the development of targeted therapies are critical in addressing the global threat posed by antibiotic-resistant staphylococci.
The presence of the PVL gene in S. aureus strains has been associated with various clinical manifestations and disease outcomes, including skin and soft tissue infections (SSTIs), osteomyelitis, and potentially malignant transformations such as PVL. Understanding the implications of the PVL gene on antibiotic resistance in S. aureus is crucial for effective clinical management and treatment strategies. Studies by Leistner et al. (2022) and Pimentel de Araujo et al. (2021) highlight the prevalence of PVL-producing S. aureus strains in clinical settings, particularly in cases of SSTIs and osteomyelitis. Leistner et al. (2022) reported a detection rate of 61.3% for the PVL gene in skin and soft tissue infections, with PVL-positive strains showing a higher recurrence rate compared to PVL-negative strains. Furthermore, PVL-SA infections are associated with specific clinical features, including leukocytolysis and dermatonecrosis, which contribute to the severity and management of these infections. The emergence of antibiotic-resistant S. aureus strains poses significant challenges in the treatment of infections. Tromp and van Strijp (2020) discussed the role of PVL and other leukocidins as virulence factors that contribute to the pathophysiology of S. aureus infections. While the exact mechanisms remain incompletely understood, the involvement of PVL in the epidemic spread and increased virulence of community-acquired methicillin-resistant S. aureus (CA-MRSA) strains underscores the need for novel treatment approaches. Effective management of PVL-associated infections requires a multidisciplinary approach, as emphasized by Khurana et al. (2021). Early and accurate diagnosis, along with appropriate intervention, is essential for therapeutic recovery and rehabilitation, particularly in vulnerable populations such as children with periventricular leukomalacia (PVL) or patients with atopic dermatitis (AD) prone to S. aureus infection. Hulme (2023) highlighted the importance of tailored treatments targeting underlying pathological mechanisms, such as SA toxins and impaired immune responses, in AD patients with S. aureus infections. Further research is needed to elucidate the complex interplay between the PVL gene, antibiotic resistance, and clinical outcomes in S. aureus infections. Studies such as those by Wei et al. (2022) and provide insights into the association between S. aureus infections and carcinogenesis, suggesting potential implications for cancer diagnosis and treatment. Additionally, ongoing efforts to characterize the molecular epidemiology of S. aureus clones causing infections, as described by Pimentel de Araujo et al. (2021), will contribute to a better understanding of disease transmission and evolution. In conclusion, the presence of the PVL gene in S. aureus strains significantly influences clinical manifestations, treatment outcomes, and antibiotic resistance profiles. A comprehensive understanding of the impact of PVL on disease pathogenesis and progression is essential for the development of targeted therapeutic interventions and infection control strategies. Collaborative research efforts combining clinical, molecular, and epidemiological approaches will further advance our knowledge of PVL-associated infections and improve patient outcomes in clinical practice.
The findings presented in this study offer valuable insights into the prevalence and association of key genetic determinants, namely the mecA gene associated with methicillin resistance and the PVL gene, among S. aureus isolates. In Figure 8A, the detection of the mecA gene is highlighted, revealing that a significant majority (82.79%) of the analyzed isolates tested positive for this genetic determinant. This high proportion underscores the pervasive nature of methicillin resistance mechanisms among the sampled S. aureus strains. Moreover, the strong association between mecA gene presence and methicillin resistance is further emphasized by its prevalent detection among MRSA isolates (67.91%). Interestingly, the observation of mecA gene detection in a subset of MRSA-negative cases (14.88%) suggests potential alternative mechanisms of methicillin resistance or discrepancies in MRSA classification. Figure 8B focuses on the detection of the PVL gene, revealing its presence in 41.86% of all analyzed isolates. Among MRSA isolates, the PVL gene was detected in 35.35% of cases, indicating its prevalence in methicillin-resistant strains. Conversely, among MRSA-negative cases, the detection rate of the PVL gene was notably lower (6.51%), suggesting a reduced prevalence among methicillin-sensitive strains. However, the significant percentage of non-MRSA isolates exhibiting the presence of the PVL gene (47.44%) indicates that it is not exclusive to MRSA strains. This highlights the potential role of the PVL gene in a broader spectrum of S. aureus infections, irrespective of methicillin resistance status. Overall, these findings underscore the complex interplay between genetic determinants, antibiotic resistance, and virulence factors in S. aureus strains. Further research is warranted to elucidate the mechanisms underlying methicillin resistance and the role of the PVL gene in S. aureus pathogenesis, which could inform the development of targeted therapeutic interventions and infection control strategies.
This study provides valuable insights into the prevalence of S. aureus infections and the associated antibiotic resistance patterns, focusing on the key resistance-associated genes mecA and PVL. The high prevalence of MRSA and MSSA underscores the importance of tailored management strategies to address these distinct variants effectively. The widespread resistance observed among S. aureus isolates highlights the urgent need for judicious antibiotic use and surveillance to mitigate antimicrobial resistance. The significant association between mecA gene presence and methicillin resistance reaffirms its pivotal role in MRSA evolution and underscores the challenges in treating infections caused by these strains. Moreover, the prevalence of the PVL gene across S. aureus strains highlights its role as a virulence factor and emphasizes its contribution to disease pathogenesis and rapid dissemination.
These findings underscore the importance of comprehensive strategies encompassing surveillance, infection control measures, and the development of novel therapeutic interventions targeting key resistance and virulence factors. Collaborative efforts across healthcare sectors are crucial to combatting antimicrobial resistance and improving patient outcomes in S. aureus infections. Further research into the molecular mechanisms underlying resistance and virulence will enhance our understanding and inform the development of targeted interventions to address this pressing public health concern.
Ethical approval for this study was obtained from the Ethics Council in Biomedical Research at Can Tho University of Medicine and Pharmacy on May 28, 2020, under Reference Number: 421/QD-DHYD. The study involving human participants strictly adhered to the ethical principles outlined in the Declaration of Helsinki. The protocol was thoroughly reviewed and approved by the Ethics Council, ensuring that all ethical requirements were met. Written informed consent was obtained from each participant prior to their involvement in the study.
To ensure transparency and reproducibility, we have deposited all the raw data underlying this study in BioStudies. You can access the data via the following link: Be-Hai Nguyen-Thi, Ngoc-Nga Pham-Thi, Hai-Yen Nguyen-Thi, Long-Nguyen Nguyen, & Trung-Son Le (2024). Unraveling the genetic basis of antibiotic resistance in Staphylococcus aureus: Focus on mecA and PVL gene. BioStudies, S-BSST1482. Retrieved from https://www.ebi.ac.uk/biostudies/studies/S-BSST1482. DOI: 10.6019/S-BSST1482.
The project contains the following underlying data:
Data are available under the terms of the CC0 1.0 UNIVERSAL (CC0).
Gratitude is extended to Can Tho University of Medicine and Pharmacy for their generous support of the research project, which included providing access to necessary samples and testing facilities. Sincere appreciation is also expressed to all those who contributed to the project and aided in the research endeavors.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
No source data required
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: molecular genetics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 27 Aug 24 |
read | read |
|
Version 1 16 Jul 24 |
read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)