Keywords
transcriptome, quantification, assembly, discovery, annotation
This article is included in the Cell & Molecular Biology gateway.
This article is included in the Nanopore Analysis gateway.
Long-read RNA sequencing technologies can produce complete or near-complete transcript sequences. Recently introduced methods for direct RNA and cDNA sequencing can provide a high-throughput strategy for the discovery of novel and rare gene isoforms. However, the high error rates in ONT sequences limit the ability to exactly pinpoint splice site boundaries when aligning reads to the genome.
In this paper, we present a novel tool called NIFFLR (Novel IsoForm Finder using Long Reads) that identifies and quantifies both known and novel isoforms using long-read RNA sequencing data. NIFFLR recovers known transcripts and assembles novel transcripts present in the data by aligning exons from a reference annotation to the long reads.
NIFFLR effectively recovers correct transcripts from simulated reads based on known transcript annotations, achieving higher sensitivity and precision compared to several previously published tools. On real data, NIFFLR shows high accuracy as measured by concordance of isoform counts to the counts computed from Illumina data for the same sample. We applied NIFFLR to a set of 92 GTEx long-read samples and produced transcript counts for both novel and known isoforms. In total, we identified and quantified 119,928 isoforms present in the RefSeq annotation of GRCh38 and 42,868 novel isoforms across 34,383 genes, more than previous studies identified in this dataset.
NIFFLR is an effective tool aimed at assembly and quantification of transcripts present in the long, high error transcriptome reads. NIFFLR is released under an open-source license (GPL 3.0) and is available on GitHub at https://github.com/alguoo314/NIFFLR/releases, and from BioConda as “nifflr”.
transcriptome, quantification, assembly, discovery, annotation
We revised the manuscript in response to the reviewers’ criticisms and in light of the development of version 2.0.0 of the NIFFLR software, as described in the updated manuscript. At the reviewers’ request, we updated Figures 3, 4, and 5 with output from the latest version of NIFFLR. Specifically, we added vertical axis labels to Figure 3 and included Pearson correlation coefficient values. We also added comparisons with Bambu in Figures 3 and 4. Figure 5 was updated to show the number of novel isoforms in GTEx long-read data, as computed by NIFFLR v2.0.0. Table 1 now reflects results from NIFFLR v2.0.0 and includes a comparison with Bambu, per Reviewer 3’s request. Additionally, we introduced a new table—now Table 2—that compares NIFFLR’s performance to other pipelines on SIRV E2 data, addressing further reviewer feedback. We rewrote the Methods section to improve clarity, as requested. In both the Introduction and Results sections, we explicitly stated that NIFFLR is best suited for well-annotated genomes, such as the human genome. A detailed list of narrative changes is provided in the point-by-point responses to the reviewers.
See the authors' detailed response to the review by Yuan Gao
See the authors' detailed response to the review by Colin Dewey
See the authors' detailed response to the review by Fairlie Reese
Direct RNA and cDNA sequencing technologies from Oxford Nanopore Technologies (ONT) produce long transcriptome reads with high yields at relatively low cost. However, the per-base error rates of ONT reads are still much higher than those of Illumina reads. Several computational tools have recently been developed to assemble transcripts and quantify isoforms in samples sequenced using ONT reads, including FLAIR (Tang AD et al., 2020), ESPRESSO (Gao Y et al., 2023), Bambu (Chen Y et al., 2023), and IsoQuant (Prjibelski AD et al., 2023). All these tools begin by mapping the long reads to the genome using the Minimap2 (Li H, 2018) aligner in spliced alignment mode. However, the high error rate of ONT reads makes it challenging to precisely identify splice sites through spliced alignment alone. Therefore, these tools incorporate additional information to locate the splice sites accurately. FLAIR can correctly identify splice sites by either using alignments of short-read RNA-seq data or by using a reference annotation. ESPRESSO accepts novel splice junctions only if at least one read aligns perfectly to the reference genome within 10 nucleotides (nt) of the splice site. IsoQuant replaces novel splice sites with nearby annotated sites within a user-defined distance and restores short, skipped exons according to the reference annotation. Bambu uses a machine-learning classifier to discover transcripts. For all these programs, misalignments can lead to incorrect identification of splice junctions, which may subsequently result in inaccurate transcript reconstruction.
Here, we present NIFFLR (Novel IsoForm Finder using Long Reads), a tool designed to construct and quantify both annotated and novel isoforms using a reference annotation and long RNA sequencing reads. Unlike other isoform identification tools, NIFFLR does not rely on a spliced aligner to map reads onto the reference genome. Instead, it extracts exons from the given annotation and aligns them directly to the long reads. NIFFLR then constructs transcripts by identifying an optimal path through the mapped exons for each long read, removes redundant transcripts that are contained within others, filters out transcripts with low read support, compares the predicted transcripts to the reference annotation, and finally quantifies both annotated and novel isoforms. For efficient exon-to-read alignment, NIFFLR uses a custom aligner based on a partial suffix array adapted from the MaSuRCA assembler (Zimin et al., 2013).
Even though NIFFLR can use exons derived from transcriptome assemblies based on Illumina data, the primary use cases for NIFFLR are genomes with high-quality annotations, such as genomes of human, mouse, and other model organisms, where almost all exons have been discovered, and alternative isoforms are, in most cases, different combinations of the existing exons. We show that, for well-annotated genomes, NIFFLR provides superior sensitivity and quantification accuracy on simulated reads, high sensitivity on the synthetic SIRV E2 dataset, and high consistency of quantification results when compared with transcript abundances computed from Illumina RNA-seq data from the same tissue.
We designed the NIFFLR algorithm to build transcripts (i.e., sequences of exons) by computing the optimal tiling of every long read using exons and transcripts provided as input. We require the following inputs: long RNA sequencing reads in FASTQ format, a reference genome sequence file in FASTA format, and a reference annotation file in GTF format. We provide a complete flowchart of the method containing the steps described below in the Supplementary Materials (Supplementary Figure 1).
First, we extract the exon sequences from the reference genome using the annotation and output them into a FASTA file. The name of each exon encodes the chromosome name, start and end position on the chromosome, the name of the gene to which the exon belongs, and its orientation. We reverse complement all exon sequences that are on the reverse strand.
We then use a version of a technique first utilized in the MaSuRCA assembler (Zimin et al., 2013) to efficiently compute approximate alignments of exons to the long reads. This alignment technique, which we refer to as psa_aligner, is based on a partial suffix array (PSA). The PSA is designed to efficiently compute approximate alignments, or alignment intervals between two sets of DNA sequences. The psa_aligner first builds a partial suffix array from a concatenated string S containing the sequences of all exons, separated by the letter ‘N’ (note that no ‘N’ characters are allowed in the reference sequence). We also record the starting position of each exon in S. Unlike a traditional suffix array, the PSA limits the suffix size to a predefined value K. The suffix array allows us to quickly locate all occurrences of a given subsequence of length K (or a K-mer) within S, and thus identify all exons and positions where a particular K-mer occurs. We then examine each K-mer in a given long read and compute all the longest common sub-sequences (LCS) of K-mers between the read and the exons, using a default value of K = 12. This value has been empirically determined to provide the best sensitivity for ONT reads with up to 10% error rates. Values of 11 or lower introduced numerous false positive alignments and slowed down NIFFLR without improving sensitivity, whereas values above 12 resulted in missed alignments of short exons. In general, the optimal value of K depends on the read error rate and the minimum exon length in the annotation. The K-mer size can be increased to 15 or 17 for lower error rate transcriptome sequencing datasets. The approximate alignment coordinates are then determined by calculating the best linear fit between the positions of K-mers belonging to the LCS in the read and on the exon. We only retain alignments where matching K-mers cover at least 35% of the bases within the match interval. Each alignment provides alignment start and end positions, along with the exon and read overhangs, as shown in Figure 1. For each exon, we record the number of K-mers in the LCS, the alignment start and end positions, and the implied start and end on the read. The implied start is calculated as alignment_start-a_overhang, and the implied alignment end is alignment_stop+b_overhang.
After building the alignments, we assign each long read to a gene locus using a “majority vote” approach. Specifically, for each read, we compute the total number of K-mers in all LCSs for all matching exons from different gene loci and assign the read to the locus L whose exons have the highest total number of matching K-mers. Alignments of any exons that belong to different gene loci are then discarded. Next, we build the transcript matching the read by finding the best tiling of the read using exons that belong to locus L. The best sequence maximizes coverage of the read while minimizing gaps or overlaps in the implied alignment coordinates. The long read defines a 5’ to 3’ forward direction, specifying a topological order. We sort the aligned exons in the order of their “alignment start” coordinates if aligned in the forward direction, or “alignment end” coordinates if aligned in the reverse direction. Since we only kept alignments of exons that all belong to a single gene locus L, the exons must all align either in the forward or reverse direction. For simplicity, below we describe the algorithm assuming all exons are aligned in the forward direction; the reverse case is treated the same, by reversing the long read.
We represent the exon tiling problem as a directed graph, where nodes correspond to exons, and node weights are exon lengths. An edge connects the 3′ end of an exon A to the 5′ end of exon B if the absolute value of the distance between their aligned positions on the read is less than 20bp. The weight of the edge between exons A and B is defined as this distance. Next, we choose the “start” nodes. A valid start node is an exon that is not connected on the 5′ end and whose 5′ end lies upstream of the read start (i.e., has a negative coordinate relative to the read origin), indicating an overhang. If no such exon exists, we select the exon(s) with the smallest positive coordinate among all exons aligned to the read. If multiple exons share the same alignment start due to alternative splicing, we use all such exons as alternative start nodes. We select “end” nodes in a similar manner: an end node is an exon not connected on the 3′ end and whose 3′ ends extends beyond the end of the read (i.e., has an overhang). If none do, we choose the exon(s) whose 3′ end coordinate is closest to the 3′ end of the read.
We solve the exon tiling problem by finding the path through the graph that starts from any start node and ends at an end node and minimizes a penalty function. The penalty is defined as the sum of edge weights along the path plus an overhang penalty, calculated as 0.1× (|5′ overhang of the start exon |+| 3′ overhang of the end exon|), where |a| denotes the absolute value of a. Ideally, there should be no gaps or overlaps between aligned exons in the transcript, resulting in a perfect path with zero weight. However, because psa_aligner computes approximate alignments, exon start and end alignment coordinates are imprecise estimates. In case of a tie, we select the path with the larger total node weight (the sum of the lengths of the exons on the path). If a read can be spanned by a single exon — either because only one exon maps to the read, or because that exon simultaneously has the start closest to the 5′ end and the end closest to the 3′ end of the read— we report that single exon as the path. If multiple exons individually span the read, we select the exon that satisfies the condition of being a valid start exon or end exon and has the smallest total overhang length. Finally, if no valid path is found, we report the exon with the longest alignment to the read. Figure 2 illustrates an example of a valid exon path. Once the best path is identified, we examine the genomic coordinates of the exons, which are encoded in their sequence IDs. We discard the path if any exons in it overlap in genomic coordinates, as this likely indicates a long read is chimeric or there is a substantial local genome rearrangement that NIFFLR cannot handle.
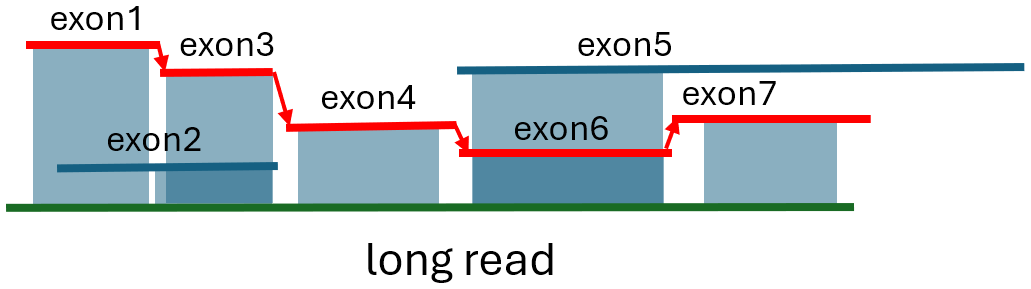
Shading shows the alignment regions. Arrows indicate links. The best path shown in red is the longest path that minimizes the gap/overlap/overhang penalty. Exon1 is chosen as the start exon because exon1+ exon3 have a longer alignment than exon2. Exon5 is alternatively spliced compared to exon6 and exon7, and its longest match is the same as exon6’s, shorter than exon6 and exon7 combined, and hence not selected for the optimal path. Exon2 is alternatively spliced as well.
We convert the best path of exons for each read into a preliminary transcript. For each preliminary transcript, we record the number of reads contributing to it, along with the average gap/overlap penalty (A mean) and the maximum gap/overlap penalty (G max) across the best path of reads that yielded the transcript. We discard all preliminary transcripts with A mean > 5 bp or G max > 15 bp. These values are empirically determined and, in general, they depend on the error rates (especially the insertion/deletion error rate) of transcriptome reads.
The next step is to examine all intron junctions in preliminary transcripts. If we find an intron junction J = (donor coordinate, acceptor coordinate) in a preliminary transcript that is not in the reference transcriptome, but there is an intron junction J′ in the reference where donor or acceptor coordinate differ by less than 10 bp, and making the adjustment to switch J to J′ in the preliminary transcript yields valid exons on the both sides of J, we make the adjustment. The rationale behind this strategy is that plus or minus 10 bp is within the error of our approximate alignment coordinates, and thus it is more likely that the actual correct junction is J′, and not J. This step results in a set of adjusted preliminary transcripts. For every such transcript, we also record all reads that yielded this transcript.
We then use the trmap tool that is part of the gfftools package to map intron chains of the adjusted preliminary transcripts to the reference transcripts provided. We only consider alignments where the intron chain of the preliminary transcript has an exact match or is contained in the reference transcript. We determine that the reference transcript is present in the sample if one of the conditions is true:
1. every intron junction in the reference transcript is spanned by at least two preliminary transcripts, or
2. one or more junctions in the reference transcript are spanned by a preliminary transcript, and that preliminary transcript does not match any other reference transcripts
3. reference transcript is intron-less, and one or more intron-less preliminary transcripts with at least five total reads in them have at least 80% (of the reference) overlap with the reference transcript
This procedure results in a list of reference transcripts that are putatively detected in the sample.
We then proceed with determining which novel transcripts are present in the sample. To do that, we apply even more stringent filtering criteria to the preliminary transcripts, discarding the transcripts with average overlap/gap Amean exceeding 2 bp and maximum overlap/gap Gmax exceeding 5bp. We then use the gffcompare tool to create a set of maximal transcripts by removing those whose intron chains are contained in longer transcripts. We call this set of transcripts “non-redundant”. Similarly to the previous step, we use the trmap tool to align intron chains of the more stringent preliminary transcript set to the nonredundant set of transcripts. We then compute how many reads span every splice junction in every non-redundant transcript. We also use the trmap tool to compare intron chains of non-redundant transcripts to the reference transcripts. Transcripts from the non-redundant set that are not contained in any reference transcript, and with all splice junctions spanned by more than two reads, are added to the set of novel transcripts. We merge the novel transcripts with the reference transcripts putatively detected in the sample to produce a final set of transcripts.
For quantification, we again use the trmap tool to compare intron chains of the preliminary transcripts to the final set of transcripts. We only use alignments that have a complete or contained intron chain match. We remind that each preliminary transcript has a number of reads N that yielded that transcript recorded. If a preliminary transcript matches M final transcripts, it contributes N/M to the count of each final transcript that it is contained in. We call N/M a distributed count. We compute read counts for each final transcript by adding distributed counts from all preliminary transcripts whose intron chains match or are contained in the final transcript. For each final transcript, we output the final read count, the intron chain, and the minimum number of reads spanning a junction, or minimum junction count. The minimum junction count can later be used in subsequent analysis filtering steps.
NIFFLR is designed to run under 64-bit Linux operating system. NIFFLR requires at least 16Gb of RAM and supports multi-core multi-threaded hardware environment. NIFFLR code consists of shell and Python scripts and C++ code. We provide installation instructions for NIFFLR on GitHub: https://github.com/alguoo314/NIFFLR. Basic usage of NIFFLR is as follows: /path/nifflr.sh -r genome.fasta -f reads.fastq -g genome.gtf.
In this section, we compare NIFFLR to other recently published methods such as FLAIR2, IsoQuant, Bambu and ESPRESSO, and discuss the results of applying NIFFLR to ONT data from the Genotype-Tissue Expression (GTEx) project (Glinos et al., 2022). We performed three evaluations to compare NIFFLR to the existing methods. First, we assessed the performance of each program on a set of simulated ONT direct RNA sequencing reads. Second, we compared the performance of the tools on SIRV E2 synthetic transcripts. Finally, we evaluated all programs on a sample from the GTEx project that was sequenced using both Illumina and ONT technologies for consistency of the transcript abundances computed from Illumina and long read data.
We simulated reads using NanoSim software (Yang et al., 2017) from the human reference genome GRCh38.p14 and its corresponding RefSeq genome annotation (RS_2024_08). We derived read error profiles from ONT reads of GTEx sample 1192X, which was sequenced with both Illumina RNA-seq and ONT technologies. We used the Illumina reads from the same sample to generate an expression profile for the simulation. Our simulated dataset contained approximately 7.8 million reads with an average error rate of 8.7% and an N50 read length of 944 bp. According to NanoSim output, the simulated set had 50,748 unique transcripts expressed.
All programs in this comparison allow the use of a reference annotation to identify and correct splice junctions, and we provided such annotation in all our experiments. Note that FLAIR and IsoQuant have options allowing them to run without annotation, but their accuracy is higher if annotation is provided. To make the evaluation more realistic, we split the reference annotation into a “core” set of transcripts, which is the set with the smallest number of transcripts where each exon was present at least once (referred to as the known set), and the rest of the transcripts (referred to as the novel set). By design, the core set contained every reference donor and acceptor splice site at least once. We provided the core set but not the novel set to all programs. This way, we ensured that some portion of the expressed transcripts were not present in the input set of the reference transcripts, enabling us to measure the programs’ ability to discover and quantify novel transcripts in addition to the known transcripts. Our simulated set consisted of reads simulated from 50,748 transcripts, of which 33,686 comprised the core set and the remaining 17,062 comprised the novel set. In our experiments, we measured the number of novel and known transcripts correctly recovered by the programs, as well as the number of false positive transcripts, using the gffcompare tool (Pertea & Pertea, 2020), to compare the transcripts’ intron chains to the reference annotation. False positives were defined as any transcripts output by the programs that did not have a complete intron chain match to a transcript in the known or novel set. Table 1 shows the comparison of the programs on the simulated data. NIFFLR has the best sensitivity in recovering all isoforms, and the best overall F1 score, while keeping the number of false positive isoforms relatively low. IsoQuant had the best precision in recovering isoforms. This result demonstrates that, when isoform discovery is the primary goal, NIFFLR is the best tool.
The best values are in bold. NIFFLR recovers the most correct isoforms total (30,349) while keeping the number of erroneous isoforms lower than FLAIR2 and ESPRESSO, resulting in the best sensitivity and F1 score for isoform recovery. IsoQuant is the most conservative and the least sensitive, both on novel and known isoform discovery. Bambu recovered the most known isoforms and the smallest number of novel isoforms.
We compared the read counts computed by each program for every transcript to the actual counts from the simulation. Figure 3a presents box-and-whisker plots of the ratios (expressed as base-2-logarithms) of the actual and computed read counts for each transcript. The box spans the upper and lower quartiles of the ratios, and the whiskers represent the range for 95% of the values, with individual outliers outside of the 95% interval shown as dots. The values of the Pearson Correlation Coefficient (PCC) between the computed and actual read counts are provided in the chart headings. The dot plots of the actual transcript counts are provided in the Supplementary Materials in Figures S2-S6. NIFFLR and IsoQuant have the best correlation of computed to the actual read counts with values of PCC of 0.998 and 0.997 respectively. Showing nearly perfect correlation. Box and whiskers show that NIFFLR has a tighter distribution than FLAIR, Bambu and ESPRESSO, though it is slightly outperformed by IsoQuant. ESPRESSO shows the worst overall performance, both in terms of the distribution’s tightness and bias. Bambu has the lowest PCC. NIFFLR detected and quantified the largest number of isoforms (30,349). Figure 3b shows a more detailed comparison of the log2 ratios between the computed and actual counts from NIFFLR and IsoQuant, for the subset of 18,686 isoforms quantified by both tools. We observe that in this comparison the accuracy is nearly identical, values of PCC are equal, with NIFFLR counts showing less overall bias. This figure suggests that the reason for the slightly lower accuracy (wider whiskers) of NIFFLR compared to IsoQuant in panel (a) is the inclusion of counts for many more isoforms by NIFFLR, capturing less reliable lower-count transcripts, which IsoQuant discards. In the simulated data comparison, NIFFLR demonstrates excellent quantification accuracy, on par with IsoQuant, and has higher sensitivity, resulting in detection and quantification of more transcripts (see Table 1).
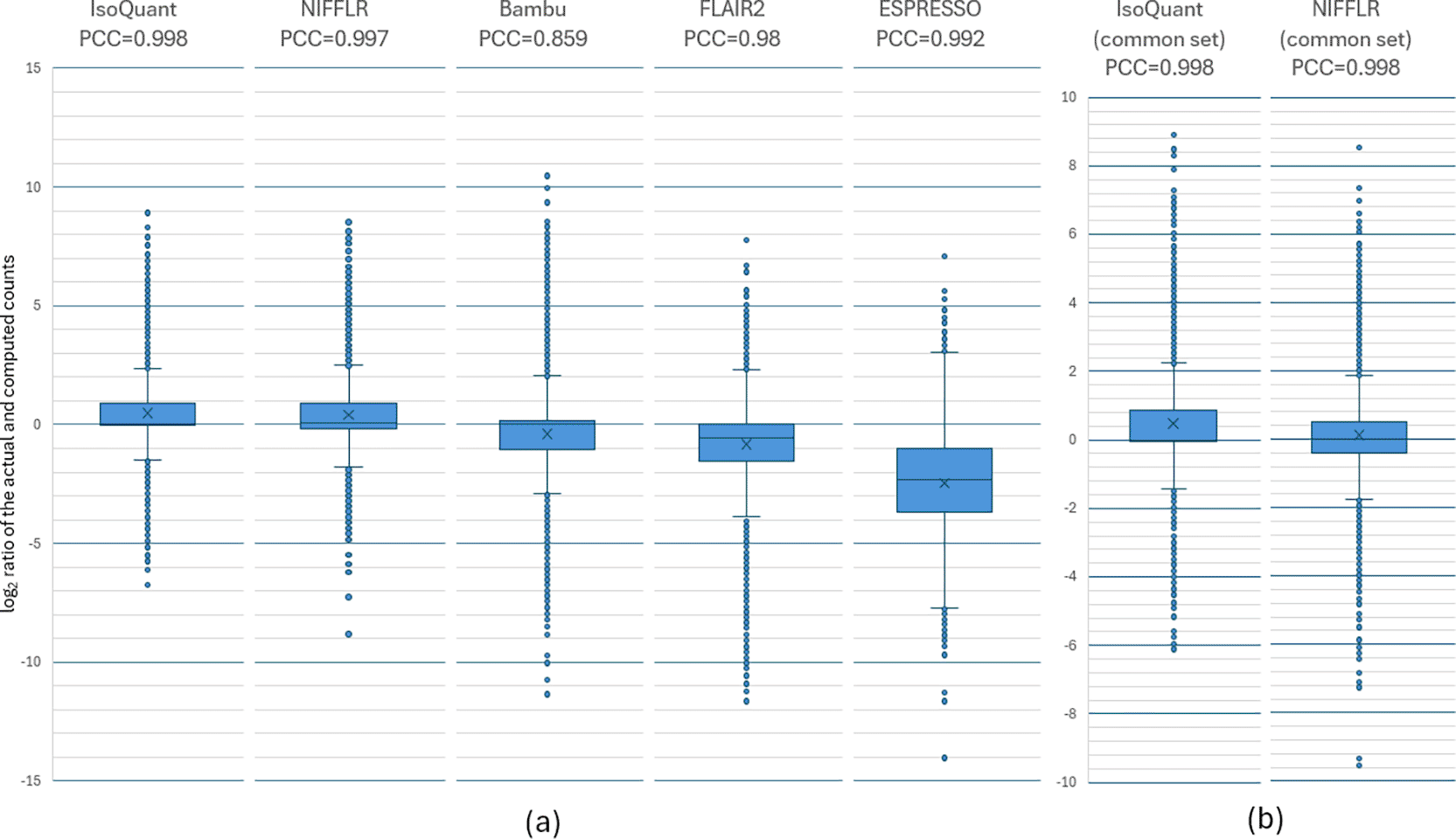
The box spans the upper and lower quartiles of the log2 ratios, and the whiskers represent 95% of the values, with individual outliers outside of the 95% interval shown as dots. The values of PCC (Pearson Correlation Coefficient) are shown above the plots. IsoQuant and NIFFLR show the least variation from the true counts in the simulated data and the highest Pearson correlation. (b) Box and whisker plots of the log2 ratios of the actual and computed read counts for each transcript from the set of 18,686 simulated transcripts quantified by both NIFFLR and IsoQuant. IsoQuant and NIFFLR show the same accuracy (the height of the box and whiskers are the same size) on this set of transcripts, however, NIFFLR counts have smaller bias (the mean and the median for NIFFLR are closer to zero). The Pearson Correlation scores are the same.
Next, we compared five programs on a SIRV-E2 sample containing 69 SIRVs (Spike-In RNA Variants). SIRVs are synthetic RNA molecules that mimic the main aspects of transcriptome complexity. They contain a variety of transcripts with alternative start sites that are nearby, as well as alternative splice junctions. We used data from the NCBI SRA experiment SRR6058583, where a mixture containing SIRV E2 mix was sequenced with the Oxford nanopore MinION sequencer with Direct RNA sequencing approach. Table 2 shows the total number of isoforms and the number of correct SIRV isoforms identified by the pipelines. NIFFLR and ESPRESSO were the most sensitive, identifying 68 of 69 isoforms present in the mixture. IsoQuant and Bambu were able to identify 65 and 51 isoforms, respectively, while outputting no extra false positive isoforms.
| # of SIRV isoforms | # of total isoforms | Sensitivity | Precision | |
|---|---|---|---|---|
| FLAIR2 | 46 | 316 | 66.7% | 14.6% |
| IsoQuant | 65 | 65 | 94.2% | 100.0% |
| NIFFLR | 68 | 89 | 98.6% | 75.3% |
| ESPRESSO | 68 | 77 | 98.6% | 88.3% |
| Bambu | 51 | 51 | 73.9% | 100.0% |
For this experiment, we selected the GTEX-1192X sample, which was sequenced with both Illumina and Oxford Nanopore instruments. The ONT data contained 7.6 million long reads with an N50 of 872 bp and a total sequence of 5.3 Gbps. In this dataset, the exact expression of existing and novel transcripts is unknown. However, we can estimate the number and abundances of the transcripts from the Illumina RNA-seq data, which provides much deeper coverage of the sample. We used StringTie2 (Kovaka et al., 2019) in reference-guided mode (using RefSeq annotation version RS_2024_08 as the reference, excluding annotations on alternative contigs and patches) to assemble transcripts from the Illumina data, and this yielded 51,827 distinct transcript variants. The reference-guided mode of StringTie does not output any novel isoforms. Table 3 shows the number of total isoforms and known isoforms found by the five long-read quantification programs when using the ONT data. NIFFLR identified and quantified 33,988 transcripts, of which 30,763 matched the reference, second to Bambu, which identified 39,985 transcripts, of which 39,413 matched the reference. Bambu was the most consistent with StringTie, identifying 27,401 transcripts present in StringTie output. To evaluate the accuracy of the quantification, we compared the read counts computed by the programs to the transcript coverage values computed by StringTie on the Illumina data from the same sample. To do the comparison in terms of read counts, we scaled the transcript coverage values computed by StringTie from the Illumina data by 1.59, the ratio of the number of bases in the Illumina reads (~8.5B bp) divided by the number of bases in the ONT reads (~5.33B bp). Figure 4 presents box-and-whisker plots of the ratios (expressed as base-2-logarithms) of the scaled transcript coverages computed with StringTie from Illumina RNA-seq reads and the read counts computed with long-read pipelines from Oxford Nanopore reads for the same sample. The box spans the upper and lower quartiles of the ratios, and the whiskers represent the range for 95% of the values, with individual outliers outside of the 95% interval shown as dots. The quantification estimates produced by NIFFLR and IsoQuant from the ONT data are the most consistent with StringTie counts derived from Illumina data for the same sample.
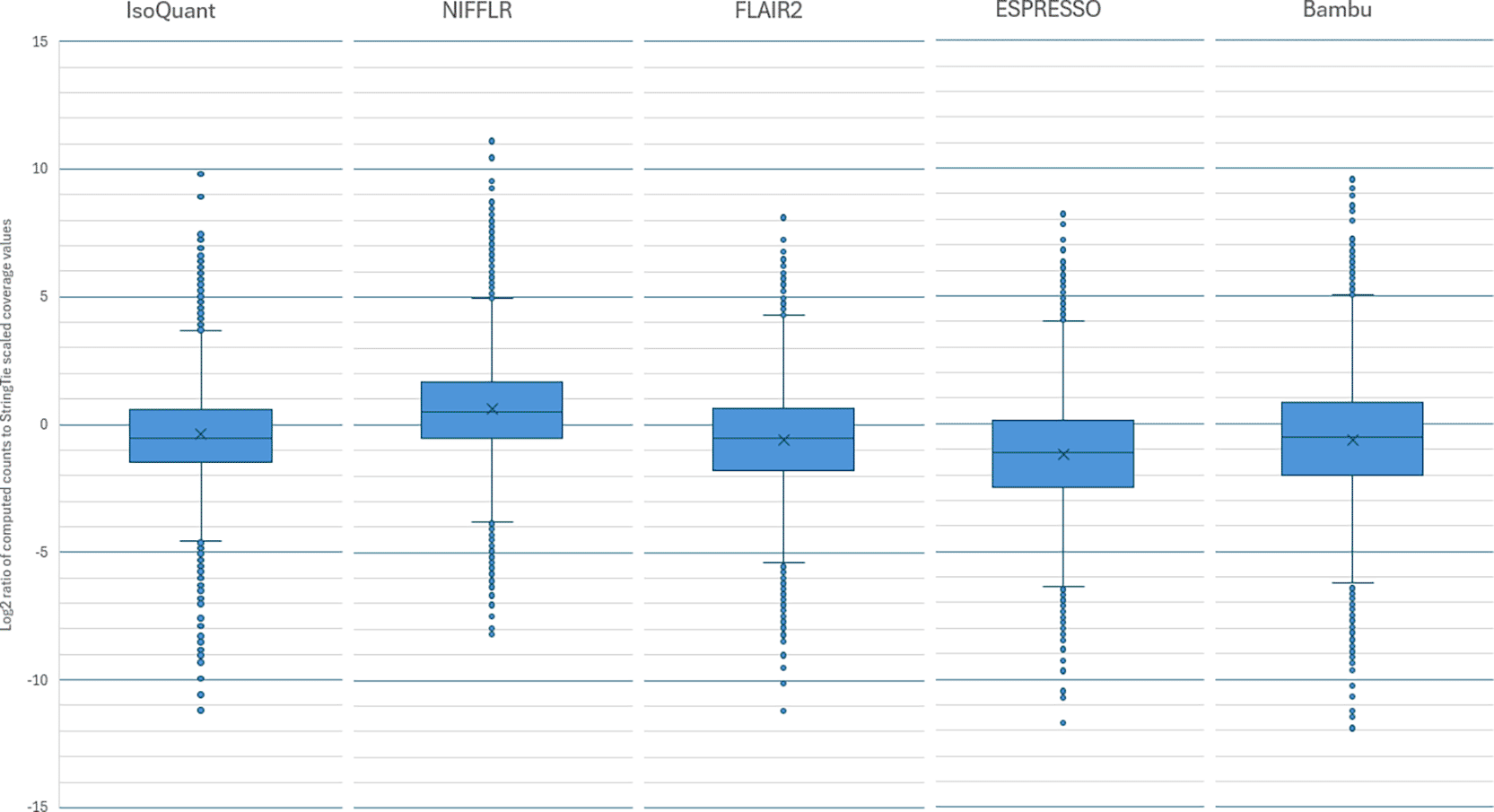
Bambu has the most isoforms in common with StringTie, followed by NIFFLR. Read counts produced by NIFFLR and IsoQuant are the most consistent with StringTie counts (smaller box and distance between whiskers).
We applied NIFFLR to identify and quantify isoforms in 92 ONT GTEx samples described in (Glinos et al., 2022), using the RefSeq annotation of GRCh38.p14 as the reference. Across all samples, we identified 119,928 known isoforms in 34,383 gene loci, and 42,868 novel isoforms in 10,487 gene loci. The number of isoforms identified by NIFFLR far exceeds the number reported by FLAIR (Glinos et al., 2022), which identified 93,718 transcripts across 21,067 genes, of which 77% were novel. 34,876 transcripts in 11,840 gene loci were in common between the set of transcripts identified by Glinos et al. (2022) and by this study. Figure 5 illustrates the distribution of counts of novel isoforms across all samples. None of the novel isoforms were present in all 92 samples, but 19 were present in all but one (91) sample. 9 of these 19 isoforms are annotated in the CHESS annotation version 3.0.1 (Varabyou et al., 2023), or in the GENCODE annotation release 47, with 3 isoforms present in both annotations. Table 4 shows the breakdown of novel and known transcripts found by NIFFLR in GTEx long-read data by tissue. As expected, the percentage of novel isoforms increases with the increase in the number of samples for a given tissue, as rare isoforms become more abundant.
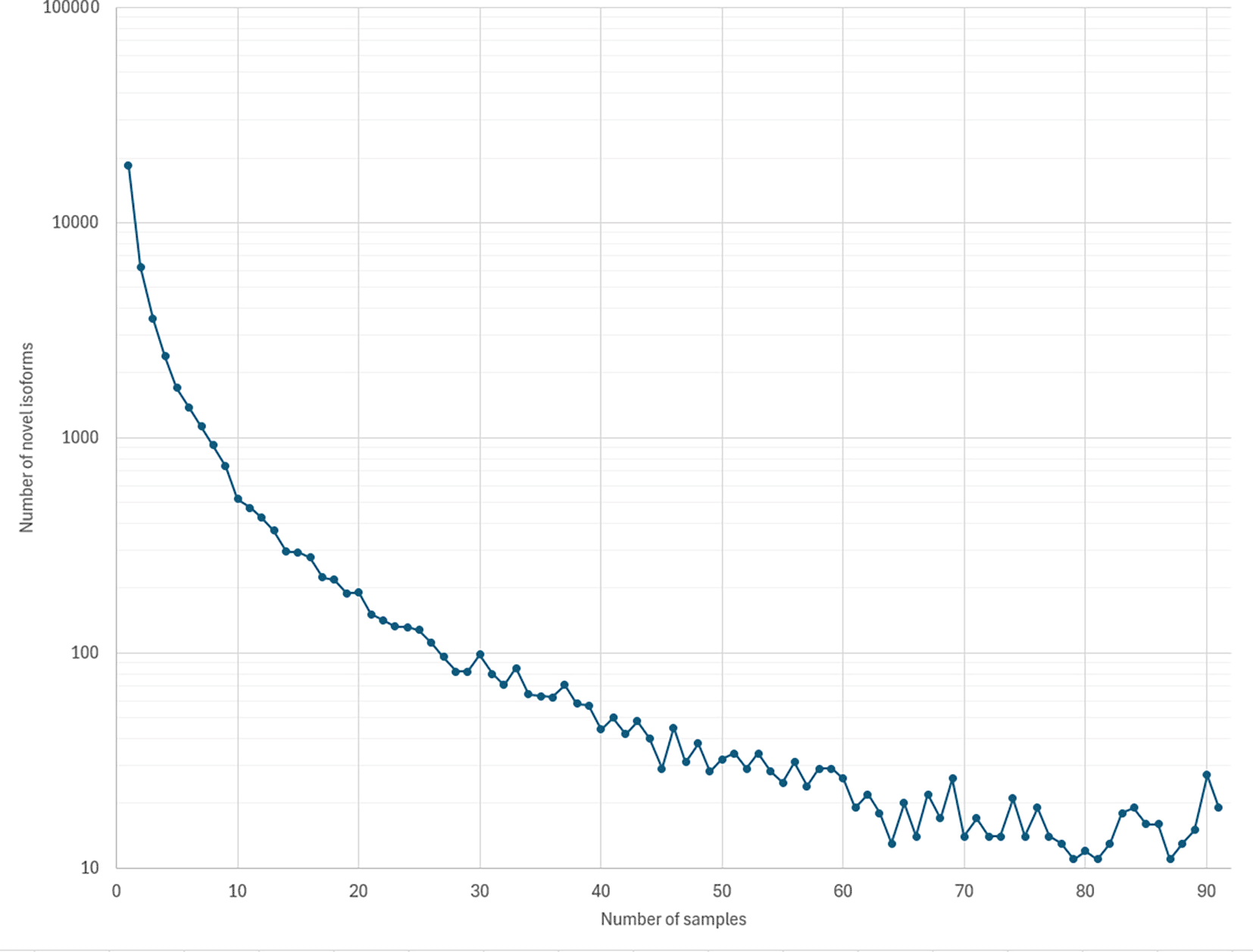
The total number of novel isoforms identified by NIFFLR in the 92 GTEX samples was 42,868. Of these, 18,337 were only seen in a single sample and 19 isoforms were identified in all 91 samples. We provide the GTF file containing all identified isoforms in the Supplementary data to this manuscript.
The share of novel isoforms increases with the increase in the number of samples for a given tissue. We used all isoforms identified by NIFFLR for the counts shown in this table.
In this manuscript, we describe a novel approach for the discovery and quantification of isoforms from long-read RNA sequencing data produced by Oxford Nanopore sequencing technology. The key difference between NIFFLR and other published programs with similar functionality is that NIFFLR aligns exons from the reference annotation directly to the reads, rather than performing spliced alignment of the reads to the genome. This approach works best for well-annotated genomes, such as the human genome, offering superior sensitivity in this case. However, NIFFLR can still be applied to genomes where their annotation is less reliable, after inferring potential exons from the Illumina RNA-seq data using transcriptome assemblers such as StringTie.
Our comparisons show that NIFFLR achieves a favorable balance of sensitivity and precision across simulated, synthetic, and real long-read datasets. On simulated data, NIFFLR recovered the largest number of isoforms overall and displayed quantification accuracy on par with IsoQuant, while surpassing other programs. These results suggest that NIFFLR is particularly well suited for studies where the discovery and quantification of novel isoforms is a primary objective.
NIFFLR is generally fast enough for research use. As shown in Table 5, NIFFLR was slower than FLAIR2, Bambu, and IsoQuant, but much faster than ESPRESSO on both simulated and real datasets. Most of the runtime for NIFFLR was spent on aligning exons to the long reads.
We ran all experiments on a 24-core Intel Xeon Gold server with 1TB of RAM, using 24 threads. Time is in hours.
| IsoQuant | FLAIR2 | NIFFLR | ESPRESSO | BAMBU | |
|---|---|---|---|---|---|
| Simulated reads | 0.7 | 1.3 | 1.9 | 45 | 1.6 |
| GTEx sample | 1.2 | 2.1 | 3.2 | 106 | 2.4 |
NIFFLR is written in shell script, Python, and C++ (the psa_aligner code). To simplify installation, we provide an install script that performs system checks and compiles all necessary executables. We have tested the installation on several popular Linux distributions including RedHat 7, 8, and 9, as well as Ubuntu 18, 20, and 22 LTS.
• The supplementary materials, transcript assembly, and quantification results computed by NIFFLR from GTEx data are available on Zenodo.
• [Zenodo]. [Supplementary information and transcripts assembled by NIFFLR software for 92 GTEx long-read transcriptome sequencing samples (revised)]. [10.5281/zenodo.17314373].
• The project contains the following underlying data: Transcripts assembled by NIFFLR software for 92 GTEx long-read transcriptome sequencing samples along with the number of samples the transcripts were observed in. Supplementary materials: commands we used to run NIFFLR and competing software for comparisons are listed in the Supplementary Information.
• gtex92.combined.gtf.gz – gzipped – GTF format file (9-column tab separated text) containing assembled transcripts on human GRCh38 assembly, chromosomes identified with NCBI RefSeq chromosome IDs.
• Supplementary materials.pdf – Supplementary materials for the manuscript titled “Assembly and quantification of transcripts from noisy long reads with NIFFLR.”
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
• Software available from: https://github.com/alguoo314/NIFFLR
• Source code available from: https://github.com/alguoo314/NIFFLR
• Archived source code at time of publication: Zenodo doi 10.5281/zenodo.17314432
• License: GNU General Public License v3.0
We thank Steven L. Salzberg, Bloomberg Distinguished Professor of Biomedical Engineering, Computer Science and Biostatistics at Johns Hopkins University for help with editing the manuscript and obtaining funding for this project.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Computational biology, bioinformatics, transcriptomics
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics, Computational Biology
References
1. Clavell-Revelles P, Reese F, Carbonell-Sala S, Degalez F, et al.: Long-read transcriptomics of a diverse human cohort reveals ancestry bias in gene annotation. Nature Communications. 2025; 16 (1). Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Long-read transcriptomics, including development of tools and their benchmarking for long-read transcriptomics analysis. I am less experienced in the field of alignment algorithms and cannot judge this implementation as thoroughly.
Is the rationale for developing the new software tool clearly explained?
Partly
Is the description of the software tool technically sound?
No
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
No
References
1. Mikheenko A, Prjibelski A, Joglekar A, Tilgner H: Sequencing of individual barcoded cDNAs using Pacific Biosciences and Oxford Nanopore Technologies reveals platform-specific error patterns. Genome Research. 2022; 32 (4): 726-737 Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics, Computational Biology
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
No
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
References
1. Pardo-Palacios F, Arzalluz-Luque A, Kondratova L, Salguero P, et al.: SQANTI3: curation of long-read transcriptomes for accurate identification of known and novel isoforms. Nature Methods. 2024; 21 (5): 793-797 Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Long-read transcriptomics, including development of tools and their benchmarking for long-read transcriptomics analysis. I am less experienced in the field of alignment algorithms and cannot judge this implementation as thoroughly.
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Computational biology, bioinformatics, transcriptomics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 03 Dec 25 |
read | read | read |
|
Version 1 20 Jun 25 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)