Keywords
Protistology, phylogenetics, phylogenomics, single-cell genomics, goniomonads, Cryptista, mitochondrial genomes
This article is included in the Genomics and Genetics gateway.
Goniomonads are heterotrophic members of a major group of eukaryotes called Cryptista and branch sister to the photosynthetic Cryptophyta. Goniomonads are poorly investigated and have recently been reclassified, dividing a single genus into ten new genera. In single-gene phylogenies, all three freshwater genera branch in a monophyletic group distinct from marine goniomonads, suggesting that a single event led to goniomonads transitioning from marine to freshwater environments. Here, we use light microscopy, scanning electron microscopy and single cell genomics to describe and determine the molecular phylogenetic placement of a new freshwater genus of goniomonad.
Light and scanning electron microscopy were used to validate the morphological characteristics of a new goniomonad. We used whole genome amplification to generate a goniomonad single-cell amplified genome. We reconstructed the phylogenetic placement of this goniomonad using single-gene and multi-gene analyses.
We characterized and established a new genus of goniomonad, named Naiadagoniomonas ponderosa n. gen. n. sp. In single-gene phylogenetic analyses, N. ponderosa branches within the freshwater clade that forms the sister to Limnogoniomonas. In multi-gene phylogenetic analyses, N. ponderosa branches with the only other representative of goniomonads available. We recovered a fragmented but high-quality single-amplified genome including a complete circular-mapping mitochondrial genome.
We conclude that diverse goniomonads are likely present in freshwater environments and single-cell genomics is an excellent tool for probing their phylogenomic diversity and mitochondrial gene content.
Protistology, phylogenetics, phylogenomics, single-cell genomics, goniomonads, Cryptista, mitochondrial genomes
The Cryptista is a clade comprising the Cryptophyta along with poorly studied lineages of heterotrophic flagellates including the Katablepharida, the Goniomonadea, and the enigmatic genus Palpitomonas, 1–6 as well as some environmental clades that include the described species Hemiarma marina.7 The three heterotrophic lineages of cryptists are inferred to be ancestrally heterotrophic, suggesting that cryptophytes independently acquired a single plastid derived from a red alga via secondary endosymbiosis.8 Goniomonads branch immediately sister to cryptophytes in phylogenomic analyses, so insights into the characters of goniomonads might help to understand the traits of the ancestral pre-plastid cryptophyte. The recently established marine goniomonad Neptunogoniomonas avonlea (Goniomonas avonlea originally) is currently the only representative of the group that has its genome sequenced.4
Goniomonads consisted of only a single genus (e.g.,9), until it was split into nine genera,10 and before a tenth genus was described in 2025.11 These recent taxonomic advances leave us with a total of three described freshwater genera and another seven from marine environments. In phylogenetic trees inferred from 18S small subunit ribosomal DNA (SSU) sequences, there is a striking divergence between a freshwater clade and all other (marine) goniomonads.3,11 This divergence suggests that there may have been only a single transition to freshwater in this lineage3 and that there may be substantial diversity of freshwater goniomonads remaining to be discovered.10–12
Here we describe a new freshwater goniomonad, which we name Naiadagoniomonas ponderosa n. gen. n. sp. We used differential interference contrast (DIC) light microscopy and scanning electron microscopy (SEM) to show that it has a typical goniomonad morphology. We sequenced the 18S SSU rDNA gene from this cell, and it branched within the freshwater clade, sister to the genus Limnogoniomonas. We isolated single cells from the culture and generated a single-cell amplified genome (SAG) using whole genome amplification (WGA). The resulting N. ponderosa SAG has a high genome completeness score. Using the SAG data, we reconstructed a phylogenomic tree which shows Naiadagoniomonas ponderosa branching sister to N. avonlea with full support. In addition, we recovered a complete circular-mapping mitochondrial genome of this species. Its mitochondrial genome has a similar coding content to the previously published genome of N. avonlea but contained genes encoding subunits of the twin arginine translocase (Tat), further inviting questions about the diversity of mitochondrial genomes in this group.
Naiadagoniomonas ponderosa was collected by scooping nearshore surface sediment and water at a creek near camp Ponderosa (34°16'58.8"N 111°08'34.6"W) in Arizona in 2022. Samples were left in the dark at ambient temperature for ~2 weeks to eliminate photosynthetic organisms. Naiadagoniomonas ponderosa was initially identified under a bright field microscope at 400x magnification. A nearly mono-eukaryotic culture has been established via sub-cultivation and single-cell isolation. All cultures were kept in the dark at 23°C in 75 cm2 culture flasks with vented caps containing 50 mL freshwater DY-V media.13 Cultures were sub-cultivated weekly in a 1:1 ratio of fresh DY-V media to original culture.
Cells were examined and recorded using a Nikon (Eclipse Ti fluorescence microscope) inverted microscope equipped with a DIC system at 1000x magnification. Images of recordings were extracted in VLC v3.20 with the snapshot feature.
Approximately 25 mL of the culture (after ~1 week of growth) was transferred to a large polystyrene Petri dish and vapor fixed with OsO4 for 10 min at room temperature. Cells were then post-fixed in 1% OsO4 for 10 min at room temperature. Fixed cells were transferred onto either a 0.2 mm or 0.4 mm Millipore membrane filters and washed in distilled water once. Fixed cells were then dehydrated using a graded ethanol series (30%, 50%, 70%, 85%, 90%, 95%) and further washed in 100% ethanol three times. Membranes containing fixed and dehydrated cells were dried with CO2 using a critical point dryer (UBC Bioimaging Facility). Dried membranes were mounted on aluminum SEM stubs and sputter coated with 2 nm Au/Pd. Stubs containing cells were viewed using a Zeiss XB350 Crossbeam scanning electron microscope (SEM) at the UBC Bioimaging Facility. This protocol is adapted from Leander and Farmer (2000).
Individual cells were manually isolated and washed 2-4 times using fresh DY-V media. Cells were placed in 0.2 mL PCR strip tubes. The REPLI-g Advance Single Cell Kit (Qiagen Cat# 150363) was used to obtain a single-cell amplified genome (SAG) following the manufacturer’s protocol. The resulting SAG was quantified using a Qubit BR DNA assay (Invitrogen Cat # Q33265). Libraries were prepared and sequenced on an Illumina NovaSeq X series instrument with 2 x 150 bp constructs (by Psomagen, Inc.).
Raw reads were quality trimmed using fastp v.0.23.2.14 Trimmed paired reads were used as an input for SPAdes version 4.0.015 and assembled under single-cell mode with default parameters, which takes multiple displacement amplification bias into account.15–17 This resulted in an unfiltered assembly (Extended Data File S5_Naiadagoniomonas_ ponderosa_assembly_dirty.fa). Once assembled, we checked and visualized SAG contamination and general assembly quality using Blobtoolkit v4.3.5.18 We used minimap2 v2.119 under default parameters to determine assembly coverage. Taxonomic affinity of each contig was determined by searching each contig into the NCBI nucleotide database using blastn v 2.16.0+20 with an e-value cutoff at 1-e50 and diamond v2.1.10.16421 into the Uniref90 database with an e-value cutoff at 1-e25. Bacterial and any obvious human contaminants were filtered out via seqkit.22 This resulted in a filtered assembly (Extended Data File S4_Naiadagoniomonas_ponderosa_assembly_clean.fa). Blobplots of the unfiltered (Extended Data Figure S1A) and filtered (Extended Data Figure S1B) assemblies show the extent of the bacterial contamination often seen in manually isolated SAGs. The completeness score for each assembly was assessed with the eukaryota_odb10 database in BUSCO 6.0.0.23 Proteins were predicted using Galba v1.24 Both the filtered and unfiltered SAG assemblies are BLAST searchable on SAGdb database at https://evocellbio.com/SAGdb/Naiadagoniomonas/.
The goniomonad SSU rDNA sequence was extracted from the SAG assembly using barrnap version 0.9 (https://github.com/tseemann/barrnap/) and aligned with a previously published data set11 using MAFFT E-INS-I version 7.481,25 manually corrected, and then trimmed using trimAl version 1.2rev5926 (-gt 0.87 -st 0.001). A Maximum-likelihood (ML) tree of the final alignment of 1,129 positions was estimated with RAxML-NG version 1.1.027 under the GTR + GAMMA model with 1,000 non-parametric bootstrap replicates.
A multi-gene phylogeny was reconstructed using the PhyloFisher dataset of cryptists with Archaeplastida as an outgroup.28 Single gene trees were manually screened and visualized via ParaSorter v1.0.4,28 for removal of paralogs, putative contaminants, and otherwise problematic sequences. Single gene alignments produced by PhyloFisher were then concatenated via Mega v12.29 Our final concatenated alignment consists of 48 taxa with 103 proteins (30,845 total aa sites). We estimated phylogeny with this concatenation under the LG+C10+F+G4 site-heterogeneous mixture model and 1,000 Ultrafast Bootstraps (UFB) using IQ-Tree2 version 2.3.6.30 The resulting tree was used as a guide tree to run a posterior mean site frequency (PMSF) analysis under the same model with 200 non-parametric bootstraps.
Using the mitochondrial genome from Neptunogoniomonas avonlea as a BLASTn query into our SAG assembly, a single contig with identical 5` and 3` sequences was identified as the circular-mapping complete mitochondrial genome of Naiadagoniomonas ponderosa. We used the MFannot31 online tool (https://megasun.bch.umontreal.ca/apps/mfannot/) to annotate the mitochondrial genomes of Naiadagoniomonas ponderosa and Neptunogoniomonas avonlea. Unidentified open reading frames (ORFs) from each mitochondrial genome were used as queries in BLAST searches into the goniomonad BLAST databases; however, no additional conserved open reading frames were identified other than those annotated by MFannot.
Naiadagoniomonas ponderosa cells were observed using DIC on a Nikon Eclipse Ti microscope. The observed cells exhibited a characteristic goniomonad morphology,32,33 with a flat oval-shape of length 6.94 μm ± 1.26 μm and width 4.60 μm ± 0.78 μm. Cells had two anterior flagella, each ~ 3.21 μm ± 0.46 μm long (0.46x cell length) and a band of visible ejectisomes near their anterior ( Figure 1A and Extended Data Video S1). Cells not attached to the surface of the cover slip swim and turn very quickly (see Extended Data Video S1, cells move into and out of field of view). Additionally, cells were observed spinning in place on the surface of the coverslip (see Extended Data Video S2).
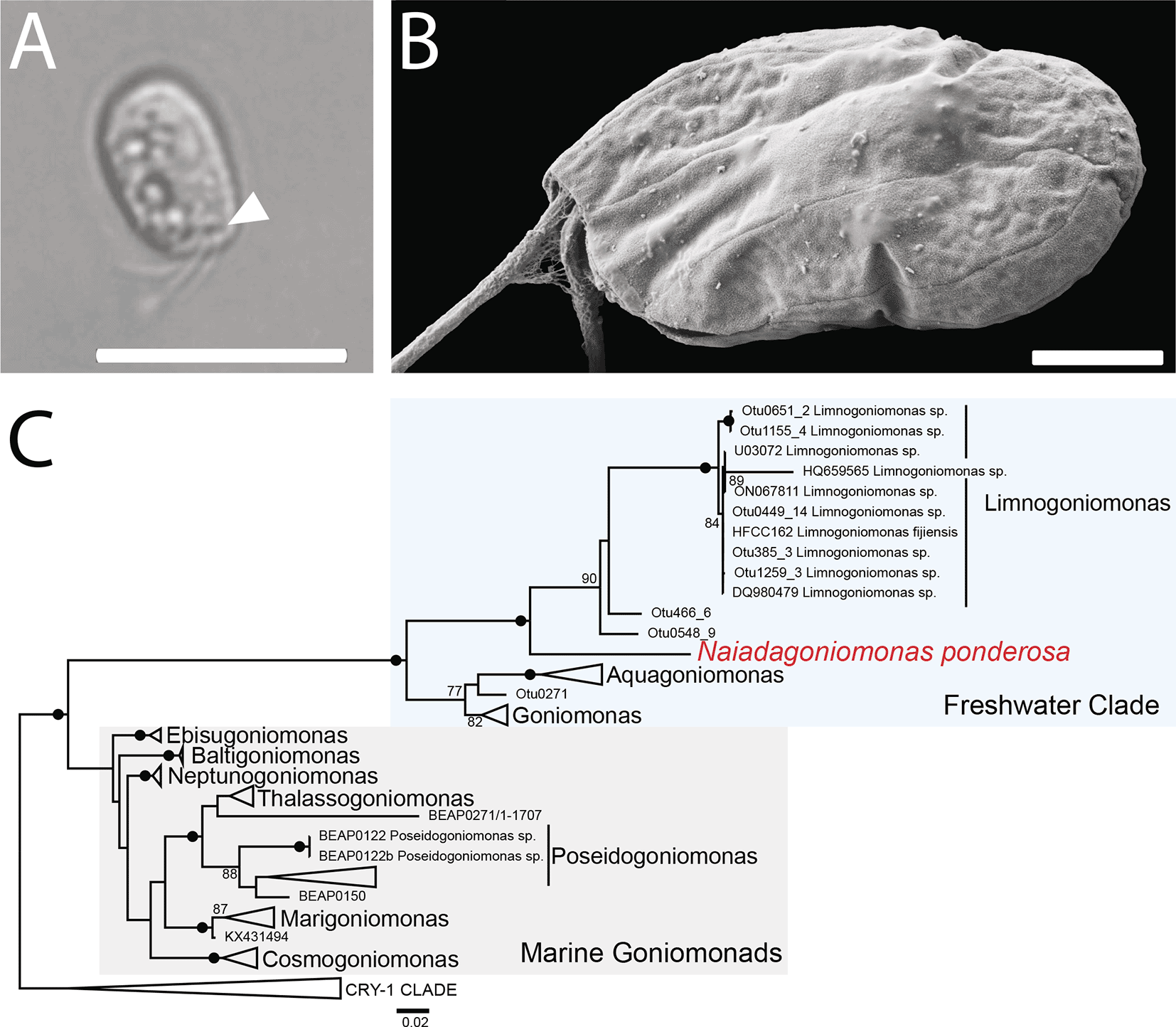
A. Differential interference contrast (DIC) micrograph showing ejectisomes (arrowhead). Scale bar = 10 μm. B. Scanning electron micrograph (SEM) of the left side of a cell showing the periplast plate pattern characteristic of this species. Scale bar = 2 μm. C. SSU rDNA phylogeny of the Cryptista showing the position of Naiadagoniomonas ponderosa. This phylogenetic tree was estimated under a maximum-likelihood framework under GTR + Gamma model and 1000 nonparametric bootstrap replicates, rooted on the CRY-1 clade as an outgroup. Bootstrap values under 75 are not shown and nodes with > 98 support are marked by a black circle.
Goniomonad periplast plate patterns have been used to differentiate marine goniomonad species from one another in tandem with molecular phylogenetic analyses.9 Scanning electron micrographs reveal three periplast plates on the left side of N. ponderosa cells ( Figure 1B), similar to the plate patterns observed in some marine goniomonad genera (i.e., Marigoniomonas) rather than the described freshwater species.10 Additional studies investigating the ultrastructure of freshwater goniomonads using SEM is necessary to compare periplast plate patterns across related species.
The N. ponderosa filtered SAG assembly constituted a total length of 256 Mbp, 170,369 contigs with an N50 of 14 kbp. To assess the level of bacterial contamination of our SAG, we used blobtools, which showed a large degree of bacterial contamination before filtering (Extended Data Figure S1A). Because our filtered SAG assembly has only a single peak that hits eukaryotes, we believe the SAG has little if any eukaryotic contamination (Extended Data Figure S1B). Furthermore, when using the Neptunogoniomonas avonlea SSU as a BLASTn query into our SAG, only a the corresponding SSU from N. ponderosa was detected, suggesting that no other eukaryotic genome was amplified. As a final check for eukaryotic contamination, we looked for contaminating mitochondrial genomes. Because eukaryotic SAGs tend to amplify mitochondrial genomes, we used the DNA sequence of the cob gene from the N. avonlea mitochondrial genome as a BLASTn query into a database containing our SAG assembly. Though this gene sequence readily hits many mitochondrial gene sequences in the NCBI nucleotide database, apart from the N. ponderosa mitochondrial genome (see below), the search recovered only a single bacterial contig. We therefore conclude that the SAG has relatively little eukaryotic contamination. The N. ponderosa SAG achieved a remarkably high BUSCO score (71.4% complete and 11.4% fragmented) compared to other SAGs (often only between 0 and 10% (see e.g.34). The published genome of N. avonlea contained more fragmented BUSCO markers but similar overall score (69% complete and 20% fragmented4). Though it is still possible that the SAG is contaminated with an unknown eukaryote, inspection of single gene trees within the PhyloFisher pipeline (see methods) suggest that most of the eukaryotic signal in this SAG is from N. ponderosa.
Next, we extracted the SSU rDNA sequence from our SAG assembly and reconstructed a single-gene phylogeny demonstrating that N. ponderosa branches within the freshwater clade of goniomonads, sister to the genus Limnogoniomonas, along with some environmental sequences ( Figure 1B). To confirm the placement of N. ponderosa in the tree of eukaryotes, we performed a phylogenomic analysis using the PhyloFisher pipeline.28 As expected, N. ponderosa branches sister to the only other goniomonad in the dataset, N. avonlea, with full support ( Figure 2). The clade comprising Cryptophyta + goniomonads was also recovered with full support ( Figure 2).
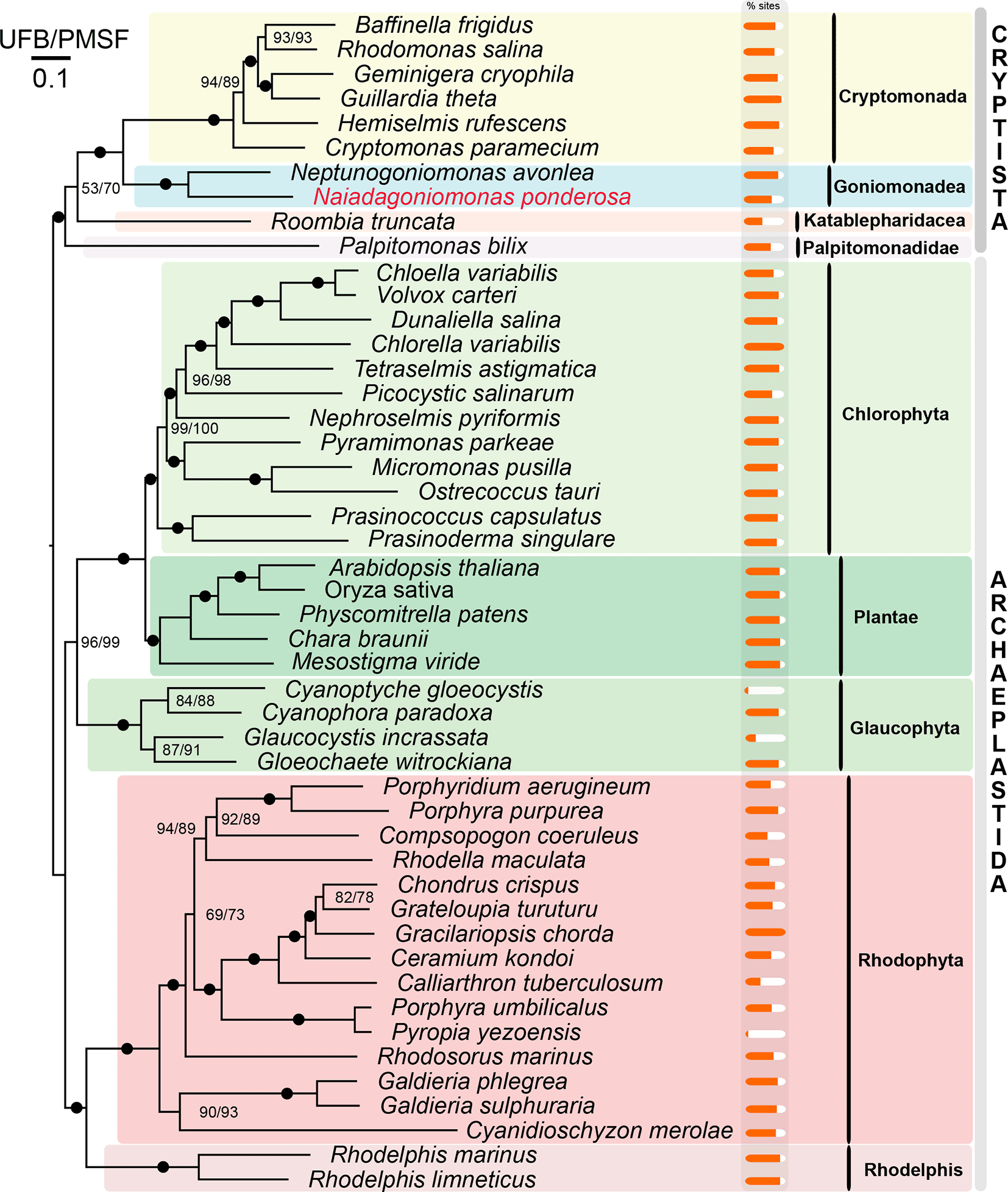
The PhyloFisher pipeline was used to estimate the phylogeny of eukaryotes using Archaeplastida as an outgroup resulting in a data matrix of 48 taxa and 103 genes (30,846 total amino acid sites). The Maximum Likelihood phylogeny was estimated using the LG+C10+F+G4 model with 1000 Ultrafast bootstraps (UFB) and 200 non-parametric bootstraps LG+C10+F+G4+posterior mean site frequency (PMSF). Naiadagoniomonas ponderosa is marked in red and bold font. Nodes with full support are marked with a black circle. Percentages of sites occupied per taxa is represented by orange bars.
In the N. ponderosa SAG, we identified a single 45,356 bp contig (30.6% GC content) representing its complete circular-mapping mitochondrial genome. The mitochondrial genome of N. ponderosa was similar in gene content compared to N. avonlea but lacked rpl10 and contained rps2 ( Figure 3). It additionally contained two protein-coding genes for the mitochondrial-encoded components of the bacteria-derived twin-arginine translocase (Tat), tatA and tatC. Though introns were identified in N. avonlea in the atp8 and cox1 genes, we identified introns in the nad2 and cox1 genes. The cox1 introns appear unrelated between the two species suggesting independent intron invasions. While rps2 is encoded in the mitochondrial genomes of katablepharids, Hemiarma marina from the CRY-1 clade,35 and cryptophytes, but not in Palpitomonas35,36 (see Figure 3). rpl10 is sporadically retained in various cryptists, though an inability to identify this gene in N. ponderosa could represent divergence beyond recognition. Meanwhile, tatA and tatC are absent in katablepharids but present in all other cryptists,35,37,38 except N. avonlea. The function of TatA and TatC in eukaryotes remains mysterious, and it is unclear why some eukaryotes retain the genes while others do not.39
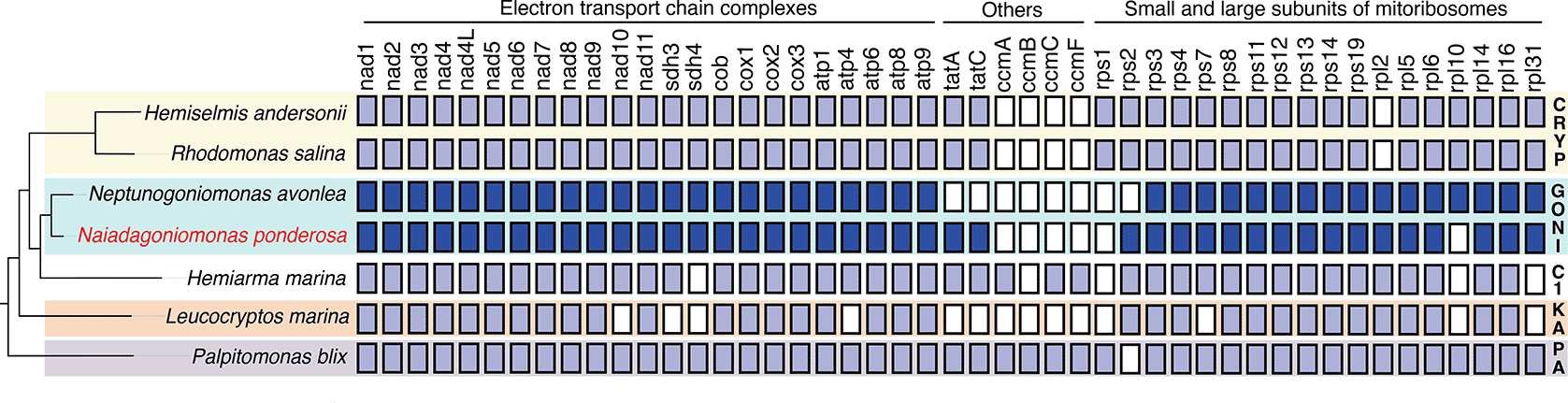
Mitochondrial genome coding contents were predicted using MFannot or mined from published literature. A schematic representation of the Cryptista phylogeny is on the left. Filled boxes represent the presence of mitochondria-encoded genes with the goniomonad genes shaded in darker blue. Abbreviations: CRYP=Cryptomonada; GONI=Goniomonadea; C1=CRY-1 clade; KA=Kathablepharidacea; PA=Palpitomonadidae.
Here, we describe a new genus and species of freshwater goniomonad Naiadagoniomonas ponderosa. Our data suggest that freshwater goniomonads have been largely overlooked and several undescribed lineages likely inhabit various freshwater habitats. We produced a high-quality SAG with high genome completeness and show that N. ponderosa branches sister to Limnogoniomonas in a single-gene tree inferred from SSU rDNA sequences and with the sole goniomonad representative Neptunogoniomonas avonlea in a phylogenomic analysis. Finally, we describe its complete mitochondrial genome and discuss differences within the cryptist clade. In conclusion, our data provide the basis for future comparative genomic investigations of goniomonads that will help to explain how goniomonads have evolved to occupy different niches in both freshwater and marine habitats.
Assignment: Naiadagoniomonas gen. nov. Rodriguez Ruiz, Severado, Cho, and Wideman (ICZN).
Taxonomic summary. Eukaryota, Cryptista, Goniomonadea, Goniomonadida.
Diagnosis. A free living, colourless, bi-flagellated heterotrophic cryptist from freshwater habitats, bacterivorous. Cell body laterally compressed, with slightly ovoid shape. Fast swimming and at times can been seen on substrate gliding and spinning under light microscopy.
Type species. Naiadagoniomonas ponderosa Rodriguez Ruiz, Severado, Cho, and Wideman
Etymology. From Naiada (Greek, singular) a freshwater nymph deity from Greece.
ZooBank Accession. LSID urn:lsid:zoobank.org:act:7AA6D9B0-1C9B-4540-BD36-3CD0E3422B7E.
Naiadagoniomonas ponderosa sp. nov. Rodriguez Ruiz, Severado, Cho, and Wideman (ICZN).
Diagnosis. Slightly ovoid flattened cell with slightly rounded posterior ~7 μm by ~ 4.5 μm and three periplast plates on the left side. Two flagella ~ 0.4x cell length.
Hapantotype: Collection of specimens on SEM stub is deposited at the Beaty Biodiversity Museum in Vancouver, British Columbia, Canada under the accession number MI-PR229.
Type habitat. Freshwater oxic sediment.
Type locality. Star Valley, Creek near Ponderosa campgrounds, Arizona. (34°16'58.8"N 111°08'34.6"W).
Etymology. From ‘ponderosa’ (english) owing to the place to which the sample was collected near Ponderosa campgrounds in Arizona.
Gene sequence. The SSU rDNA sequence extracted from the single-cell genome assembly of N. ponderosa is deposited under GenBank Accession number PX375661.
ZooBank Accession. urn:lsid:zoobank.org:pub:6301F368-B4AA-4D99-B8B3-387EFEFF5074
Raw reads are available in the NCBI sequence read archive (SRA): PRJNA1330332
Extended Data are available at FigShare under CC-BY 4.0.
Phylogenomic placement, description, and mitochondrial genome of a novel freshwater goniomonad from Arizona: Naiadagoniomonas ponderosa n. gen. n. sp.
DOI: https://doi.org/10.6084/m9.figshare.30153610.v140
The project contains the following underlying data:
Extended Data Figure S1 Blobplots.pdf
Extended Data File S1_Naiadagoniomonas_complete_mtDNA.fasta
Extended Data File S2_Neptunogoniomonas_avonlea_mfannot.new
Extended Data File S3_Naiadagoniomonas_ponderosa_mfannot.new
Extended Data File S4_Naiadagoniomonas_ponderosa_assembly_clean.fa
Extended Data File S5_Naiadagoniomonas_ponderosa_assembly_dirty.fa
Extended Data Video S1_Naiadagoniomonas_ponderosa.mp4
Extended Data Video S2_Naiadagoniomonas_ponderosa.mp4
We thank Statton Tinker for collecting the sample, and Gordon Lax, Nicholas Fry, Alexander Tice, and Robert Jones for bioinformatic advice.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)