Keywords
Fibrosarcoma, HT1080, SP600125, CIP2A, JNK2, siRNA, PP2A
Fibrosarcoma, HT1080, SP600125, CIP2A, JNK2, siRNA, PP2A
The manuscript has now been revised based on the comments received from the respectable reviewer. New data showing additional controls, along with the specific explanation to clarify the siRNA based specificity issues, has been now added. Notably, an additional small siRNA screen involving kinases previously demonstrated to be sensitive to SP600125 has been carried out, and resulted in identification of CHK1 as the kinase positively regulating CIP2A expression in HT1080 cells. This is in accordance to recent observations seen in gastric, prostate, breast and cervical cancer cells (Khanna et al., Cancer Research 2013). Additionally, the requested p-values and text have been modified to accommodate and explain the new findings.
See the author's detailed response to the review by Kristopher Clark
It has been recently established that regardless of phenotypic variability between different cancer types, perturbation of a limited number of genetic elements is sufficient to induce transformation in different human cell types1. Experimentally, it was demonstrated that activation of RAS and telomerase (TERT), along with inactivation of the tumour suppressor proteins P53 and Retinoblastoma protein (RB) can immortalize a variety of human cell types, which can subsequently transform to a tumourigenic state in response to inhibition of protein phosphatase 2A (PP2A)1,2. Various independent studies have shown that inhibition of PP2A activity is a pre-requisite for human cell transformation1,3–5. Therefore, understanding the mechanisms by which PP2A is inhibited in cancer cells is vital for developing new anti-cancer therapies.
Cancerous Inhibitor of Protein Phosphatase 2A (CIP2A) is a recently identified oncogene, which has been demonstrated to inhibit the endogenous tumour suppressive activity of PP2A in cancer cells6. Several layers of evidence, both from us and others, have shown CIP2A to be required for malignant cell growth and in vivo tumour formation6. In addition, the prognostic role of CIP2A has been demonstrated in several human tumours6. Moreover, since CIP2A overexpression has been observed at a high frequency in most human cancers studied so far6,7, identification of mechanisms regulating its expression in human cancers becomes important to address.
Although several transcription factors like MYC8, ETS19, E2F110 and ATF211 have been identified as positive regulators of CIP2A in various carcinomas, factors influencing CIP2A expression in non-hematopoietic mesenchymal cells or sarcomas, are yet to be discovered. Notably, CIP2A amplification has been observed in soft-tissue sarcomas12. In addition, since CIP2A was identified using HT1080 (human fibrosarcoma cell line) cell extracts13 this cell line was selected to dissect the mechanisms for high CIP2A expression in sarcomas. Since p38, ERK and JNK signalling pathways are commonly perturbed in cancers, we assessed the role of these pathways in CIP2A expression in HT1080 cells. To this end, respective small molecule kinase inhibitors, namely SB203580 (p38 pathway inhibitor), PD98059 (MEK pathway inhibitor) and SP600125 (JNK pathway inhibitor) were used to inhibit signalling through these pathways in HT1080 cells.
SP600125 was purchased from Calbiochem (Cat No. - 420119, Merck-Millipore CAS 129-56-6, San Diego, CA) and stocked as a 20 mM solution in DMSO. PD98059 was purchased from Calbiochem (Cat No. - 513000, Merck-Millipore, San Diego, CA) and stocked as 40 mM stock in DMSO. SB203580 was purchased from Calbiochem (Cat No. - 559389, Merck-Millipore, San Diego, CA) and stocked as 20 mM.
The siRNAs to inhibit CIP2A expression were obtained from Eurofins MWG operon (Ebersberg, Germany). Either of the following double-stranded oligonucleotides was transiently transfected into HT1080 cell line as CIP2A siRNAs: CIP2A.1, 5´-CUGUGGUUGUGUUUGCACUTT-3´, and CIP2A.2, 5´-ACCAUUGAUAUCCUUAGAATT-3´. As a control, a scrambled siRNA with the sequence 5´-UAACAAUGAGAGCACGGCTT-3´ was used instead. HP-validated siRNAs for human JNK1 and JNK2 were purchased from Qiagen Sciences (Germantown, MD). Either of the following oligonucleotides were transiently transfected into HT1080 at 30%–50% confluency in a six-well plate were transfected with the siRNA in antibiotic free medium using RNAiMAX Reagent (Invitrogen, Carlsbad, CA), according to the manufacturer’s instructions.
Proteins were extracted in hot Laemmli sample buffer and subjected to immunoblot analysis. Thirty micrograms of total protein extracts was separated by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (Bio-Rad Laboratories, Helsinki, Finland) and transferred to nitrocellulose membranes (Thermo Scientific Pierce Protein Biology Products, Rockford, USA). Membranes were blocked with 5% nonfat milk in Tris-buffered saline (TBS; 20 mM Trizma Base and 150 mM NaCl dissolved in distilled water and adjusted with HCl to pH 7.5) containing 0.1%-NP40 (Igepal Ca-630; Sigma-Aldrich)8. Nitrocellulose membranes (Thermo Scientific Pierce Protein Biology Products, Rockford, USA) were incubated with antibodies to JNK1 (Cat. No. sc-1648: 1:500 dilution, Santa Cruz Biotechnology, Santa Cruz, CA), JNK2 (Cat. No. sc-827:1:500 dilution, Santa Cruz Biotechnology, Santa Cruz, CA) in 5% milk in TBS-NP40 (Igepal Ca-630; Sigma-Aldrich) at 4°C overnight, with a 1:5000 dilution of the rabbit polyclonal anti-CIP2A antibody8 at 4°C overnight, or with a 1:1000 dilution of goat polyclonal anti-β-Actin antibody (Cat. No. sc-47778, Santa Cruz Biotechnology) at room temperature for 1 hour.
HT1080 cells originally were obtained from ATCC and were cultured in DMEM (Gibco) supplemented with 10% (v/v) fetal calf serum (FCS), 2 mM glutamine, 100 units/ml penicillin and 100 µg/ml streptomycin (Bio-Whittaker Europe, Verviers, Belgium).
Total mRNA was extracted from cells using the RNeasy kit (Qiagen, Valencia, CA) and converted to cDNA by using the M-MLV Reverse Transcriptase, RNase H Minus, Point Mutant cDNA synthesis kit (Promega Corporation, Madison, WI). cDNAs were subjected to quantitative real-time polymerase chain reaction (PCR) by using the Light Cycler (Roche Diagnostics, Mannheim, Germany) and SYBR Green PCR Master Mix kit (Roche Diagnostics). Primer sequences (Sigma-Proligo, St Louis, MO) used for PCR of CIP2A were as follows: CIP2A forward, 5´-CTGGTGAGATAATCAGCAATTT-3´ and CIP2A reverse, 5´-CGAAACATTCATCAGACTTTTCA-3´. Transcript levels were normalized to levels of TATA-binding protein (TBP) or β-Actin expression, which were determined by PCR of the same samples using the following primers: TBP forward, 5´-GAATATAATCCCAAGCGGTTTG-3´, and TBP reverse, 5´-ACTTCACATCACAGCTCCCC-3´; Actin forward, 5´-CGAGCACAGAGCCTCGCCTTTGC-3´, and Actin reverse: 5´-CATAGGAATCCTTCTGACCCATG-3´.
Cancer cell line encyclopaedia (http://www.broadinstitute.org/software/cprg/?q=node/11) was used to get the expression levels of JNK1 and JNK2 in HT1080 cells14. This is a resource which provides analysis and visualization of DNA copy number, mRNA expression, mutation data and more, for 1000 cancer cell lines.
To determine the oncogenic signalling pathways that maybe involved in regulating CIP2A expression in human fibrosarcoma, HT1080 cells were treated with small molecule inhibitors for the p38 (SB203580; 20 µM), JNK (SP600125; 10 µM) and ERK (PD98058; 20 µM) signalling pathways. As previously observed in gastric cancer cells9, PD98058 reduced CIP2A mRNA and protein expression in HT1080 cells (Figure 1A and B). Importantly, while SB203580 showed negligible effect, SP600125 potently inhibited CIP2A mRNA and protein expression in HT1080 (Figure 1A and B) cells. Furthermore, inhibition of CIP2A protein expression by SP600125 was observed to be both time and concentration dependent (Figure 1C and D).

A. qRT-PCR showing the effect of small molecule inhibitors against the p38 (SB203580), MEK-ERK (PD98059) and JNK (SP600125) pathways on CIP2A mRNA expression (12 h timepoint; Shown is mean values + S.D., or representative results from three independent experiments. Student T-test was used obtain the statistical significance value) B. Western blot showing the effect of small molecule inhibitors against the p38 (SB203580), MEK-ERK (PD98059) and JNK (SP600125) pathways on CIP2A protein expression (24 h timepoint). Shown is the representative picture of two independent experiments. C and D. Effect of small molecule inhibitors against JNK (SP600125) pathway on CIP2A protein expression in concentration (C) and time (D) dependent manner. Shown is the representative picture of two independent experiments.
SP600125-mediated inhibition of CIP2A expression suggested the involvement of the c-Jun N-Terminal Kinases (JNKs) in regulation of CIP2A expression in HT1080 cells. Since the JNK3 isoform is well known to be neural-specific15, specific validated siRNAs against JNK1 and JNK2 isoforms were transfected into HT1080 cells and CIP2A expression estimated. Although, both JNK1 and JNK2 siRNAs reduced their target proteins (JNK1 and JNK2 respectively), there was no change in CIP2A protein expression with either of them individually (Figure 2A,B and Supplementary Figure 1A) or in combination (Figure 2C). Since the JNK2 isoform is expressed more than 10-fold higher than the JNK1 isoform in the HT108014 (Figure 2D) cell line we transfected two validated and specific siRNAs against JNK2. In addition, we also transfected the HT1080 cells with two different CIP2A siRNA as positive controls. In line with our previous observation (Figure 2B and C), the two different siRNAs for JNK2 knocked out JNK2 expression (Figure 2E), while CIP2A expression remained unaltered. Surprisingly, two different CIP2A siRNA, which worked well as positive controls, efficiently decreased JNK2 expression (Figure 2E). SP600125 has been demonstrated to inhibit various kinases like CDK2, DYRK1 and AMPK even more effectively than JNK kinase itself16 (Supplementary Table 1). Interestingly, recently CHK1 kinase (also inhibited by SP600125; Supplementary Table 1) was shown to inhibit CIP2A levels both in vitro and in vivo6,17,18. Therefore, we transfected HT1080 cells with 2 different siRNAs against CHK1 (Kinase almost as sensitive to SP600125 as JNK) and with siRNAs against CDK2 and DYRK1 (two other kinases that are more sensitive to SP600125 than JNK). As shown in Supplementary Figure 1B, two different siRNAs against CHK1 decreased CIP2A expression n HT1080 cells.
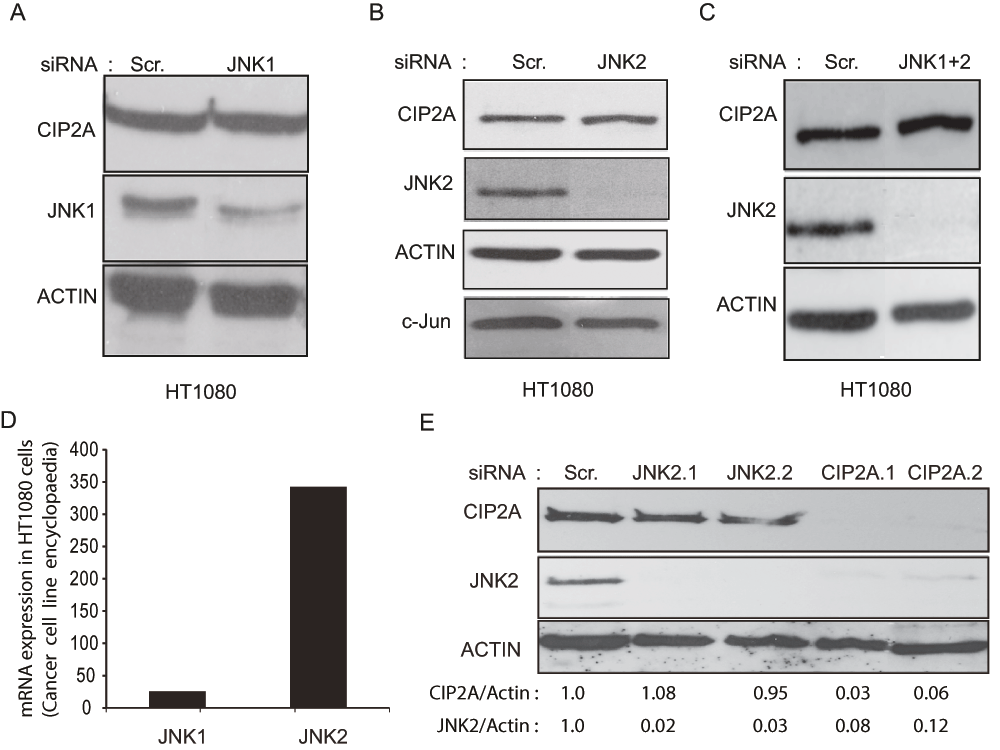
A, B, and C. Western blots showing the effect of JNK1 (A), JNK2 (B) and combination of both JNK1 and JNK2 (C) siRNAs on CIP2A protein expression, 72 h post-transfection. Shown is a representative result from two independent experiments. D. mRNA expression of JNK1 and JNK2 in HT1080 cell line from the cancer cell encyclopaedia study. E. Western blot showing the effect of two different siRNAs specific for JNK2 and CIP2A proteins and their protein expression levels 72 h post-transfection. The numbers below the blot are the quantified values for CIP2A and JNK2 protein levels normalized to Actin protein levels, relative to the levels in Scrambled (control) transfected cells. Shown is a representative result of two independent experiments.
Even though small molecule inhibitors are an emerging therapeutic option against cancers, the specificity issues limit their potential to be used in clinics. They have been extensively used to study various cell signalling pathways. In particular SP600125 has been used to study the effect of C-Jun N-Terminal Kinases (JNKs) in various processes19–21. Our results suggest that even though we were able to see decrease in CIP2A expression in HT1080 cells on treatment with SP600125 (Figure 1), at doses used previously to inhibit c-Jun NH2-terminal kinase (JNKs) activity22,23, we were not able to validate the findings using two different siRNAs specific to the JNK2 kinase (Figure 2E). Interestingly, a decade ago a previous study emphatically demonstrated the effect of SP600125 on different kinases16. The study revealed that SP600125, although a JNK inhibitor, could inhibit the activity of several other kinases like CDK2, DYRK1 and CHK116. Notably, we here identify CHK1 as the potential SP600125 sensitive kinase that positively regulates CIP2A expression in HT1080 cells (Supplementary Figure 1B).
Surprisingly, two different CIP2A siRNAs used as positive controls, decreased JNK2 expression levels in HT1080. This has also been observed in a separate study in an epithelial origin cell line, HeLa24. Interestingly, JNK2 has been shown to regulate CIP2A expression via ATF2 transcription factor in mouse embryo fibroblasts (MEFs)11 in the pre-transformed stage. Since we observe the vice versa in fully transformed HT1080 cells, it can suggest that there may be a molecular switch between JNK2 and CIP2A which may have a possible role in the RAS-transformation of mesenchymal cells. Nevertheless, the functional consequence of CIP2A-mediated JNK2 expression in mesenchymal cells would require further exploration.
Altogether, our study highlights the need for the validation of results obtained by small molecule treatments with independent approaches like two or more target specific siRNAs, shRNAs or use of inducible systems like RNAi or Tamoxifen/Tetracycline-induced overexpression systems8.
The work was funded by Academy of Finland through the Tampere Graduate Program in Biomedicine and Biotechnology.
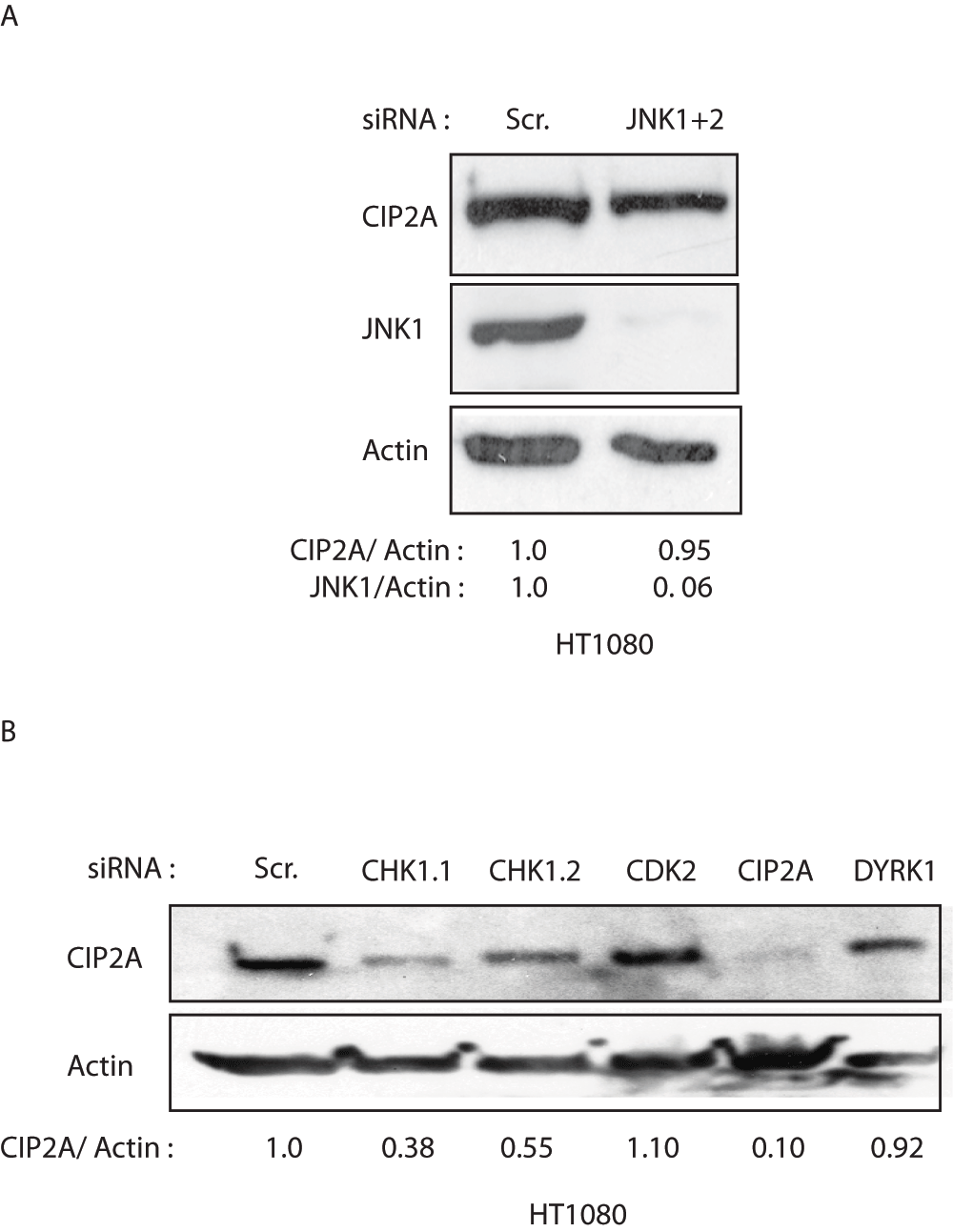
A. Western blot showing JNK1, CIP2A and Actin protein levels in HT1080 cells transfected with either control (Scr.) or combination of siRNAs against JNK1 and JNK2. The numbers below the western blot indicate the quantification of CIP2A and JNK1 protein levels after normalization to Actin levels. B. Western blot showing CIP2A and Actin levels in HT1080 cells transfected with siRNAs indicated for specific kinases previously known to be sensitive to SP600125 chemical inhibitor. The numbers below the western blot indicate the quantification of CIP2A protein levels after normalization to Actin levels.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 11 Dec 13 |
read | read |
|
Version 1 14 Aug 13 |
read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)