Keywords
DNA, mRNA, ribosomes
DNA, mRNA, ribosomes
Class 1 integrons are capable of inserting, excising and rearranging gene cassettes by a site-specific recombination mechanism. These assembly platforms can also act as expression systems due to the presence of a promoter region (Pc), which drives the expression of genes captured by integron1. Moreover, naturally occurring integrons may have a second promoter (P2), which is activated by the insertion of three G residues between -35 and -10 hexamers1. Gene cassettes are generally promoterless units associated with a recombination site (attC or 59-be), which confers the ability of each structure to be mobilized independently2. In addition, attC sites regulate the translation of downstream cassettes due to their peculiar sequences composed by imperfect inverted repeats. The formation of stem-loop structures by attC sites prevents ribosome progression throughout mRNA, reflecting in a decreased expression of more distal genes regarding Pc3. Although rare, fused cassettes may be generated by partial or total loss of the first attC, retaining both complete coding regions and, therefore, creating permanent gene arrays comparable to bacterial operons4. The functionality of such structures has been indirectly inferred by the resistance profile of transformants carrying the fusion5; however, the transcription itself has never been verified.
This study showed the dynamics of fused cassette mobilization, the co-transcription of the gcu14-bla GES-1/aacA4 cassette array and the effect of cassette position on transcription levels in Pseudomonas aeruginosa wild lineages carrying class 1 integrons. Moreover, the presence of translation signals in this gene cassette array was determined.
An unknown Open Reading Frame (ORF), gcu14, followed by the fused cassette blaGES-1/aacA4, created by partial loss of GES-1 attC were present in integrons from clinical P. aeruginosa isolates (PS1 and PS26)6. Total RNA was extracted and purified according to the manufacturers instructions with the SV 96 Total RNA Isolation System (Promega). Northern blot using 7 μg of total RNA from PS1 and PS26 was performed in order to detect the transcript originated from gcu14-blaGES-1/aacA4 cassette array. After electrophoresis in a denaturing-formaldehyde 1.5% agarose gel, the total RNA was transferred to the Hybond-N+ nylon membrane (GE Healthcare) by upward capillary transfer. An amplicon of 519bp corresponding to part of the blaGES-1 gene was used as a probe (Table 1) in hybridization assay. The GES probe was labeled with the AlkPhos Direct Labelling kit (GE Healthcare) and hybridized with the target RNA immobilized on the Hybond-N+ membrane as recommended. The chemiluminescence was detected with the CDP-Star detection reagent (GE Healthcare) according to manufactures. Immediately after applying the detection reagents, the blot was drained, incubated five minutes at room temperature and exposed to the Hyperfilm ECL (GE Healthcare) for 60 minutes at room temperature.
| Primers for conventional PCR | |||
|---|---|---|---|
| Primer | Primer sequence (5' – 3') | Size (bp) | Target |
| Ges F Ges R | GCGTTTTGCAATGTGCTC CCAGTTTTCTCTCCAACAACC | 519 | Internal fragment of blaGES gene |
| Primers for inverse PCRa | |||
| Gcu14 FSQ Gcu14 RSQ | AGCAATCAACACACAGGGG CTCGCGTAAATGCACCGCTT | 130 | gcu14 circular form |
| GES FSQ GES RSQ | CAAGTTATTACACAACTCAT AGTGCGTGAATGAAGCGCAT | 110 | blaGES-1 circular form |
| AACA4 FSQ AACA4 RSQ | GCCAGGCATTCGAGCGAACAC ATTTAGCCACTCACATAGAGC | 188 | aacA4 circular form |
| Primers and probes for real time PCR (TaqMan) | |||
| RpsL F RpsL R Probe | GCCTGCGCTGCAAAACT TTTCGGCGTGGTGGTGTAT TCGTGGCGTATGCACC | 67 | rpsL transcripts |
| Gcu14 F Gcu14 R Probe | CATGCGCTTCTTGGTTCGT ACGCCAGCTTGGATGCAA ATGCCACGAGACCTT | 56 | gcu14 transcripts |
| Ges F2 Ges R2 Probe | GTGCAGCTTAGCGACAATGG CACAGAGTCGCCAATTTTACGA AATTGCAGCAGGTCCGCC | 99 | blaGES-1 transcripts |
| AacA4 F AacA4 R Probe | CAAGCGTTTTAGCGCAAGAGT TCGGCTCTCCATTCAGCATTG CCGTCACTCCATACATTG | 59 | aacA4 transcripts |
| Ges F3 AacA4 R2 Probe | TCCTGAGCACGGACAAATAG TCATAGAGCATCGCAAGGTC TTCCGTCACACTGCGCCTCA | 134 | Polycistronic transcripts |
In order to verify whether the relative position of gene cassettes on the variable region plays a role in transcription level, real-time RT-PCR reactions using the TaqMan System (Applied Biosystems) were performed with primers and probes detailed in Table 1. The single-copy ribosomal rpsL gene of the P. aeruginosa chromosome was amplified by PCR (Table 1) and used as a reference gene for normalization. The relative quantification (RQ) results were presented as ratios of gene transcription between the target gene (cassettes) and the reference gene (rpsL), which were obtained according to the following equation: RQ=2-ΔCT, where CT is the value corresponding to the crossing point of the amplification curve with the threshold and ΔCT=CT target gene minus CT reference gene. The effect of cassette position on gene transcription was considered significant when the ratios obtained between RQ values (RQ value of cassette 1/RQ value of cassette 2) were ≥2.0, taking into account the standard deviation intervals.
In order to induce cassette excision from integrons, PS1 and PS26 strains6 were submitted to thermal stress during the log growth phase to induce integrase activity. Cells were grown on Luria-Bertani (LB) broth medium (OXOID) at 37ºC for two hours. subsequently, the bacterial cultures were submitted to a heat shock at 4ºC for 30 minutes and immediately incubated at 42ºC for another 30 minutes. Briefly, the total DNA from PS1 and PS26 cultured under thermal stress were obtained with the Wizard Genomic DNA purification kit (Promega) following manufacturer recommendations and used as templates in inverse PCR reactions. The inverse PCR was performed with primers facing outwards towards the ends of gcu14, blaGES-1 and aacA4 so that only circular gene cassette configurations would be amplified. The reactions target the circular forms of gcu14, blaGES-1, aacA4 and blaGES-1/aacA4 fusion by using the primers and combinations described in Table 1. The inverse PCR was performed using Platinum Taq DNA Polymerase reagents (Invitrogen), and the following components were added to a sterile 0.2-mL tube: 5 µL of 10X PCR buffer (1X final concentration); 1 µL of 10mM dNTP mixture (0.2 mM each); 1.5 µL of 50mM MgCl2 (1.5 mM final concentration); 2 µL of 15 µM of each primer (30 µM each); 100 ng of template DNA; 0.3 µL of Platinum Taq DNA Polymerase (1U final concentration). The tubes were incubated in the Eppendorf MasterCycler (Eppendorf) at 94°C for 2 minutes and PCR amplification was performed in 40 cycles consisting of: 94°C for 30 seconds; 55 °C for 30 seconds; and 72°C for 3 minutes. The amplicons generated with the inverse PCR were purified using Wizard SV Gel and PCR Clean-Up system kit (Promega) and directly sequenced on both strands. Sequencing reactions were performed with Big Dye Terminator RR Mix (Applied Biosystems) in an ABI 3730 XL DNA Analyzer (Applied Biosystems). Nucleotide sequences were compared to those available in the GenBank database accessible on the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). All primers used in PCR, sequencing and real time RT-PCR are described in Table 1.
Analyses in silico were performed to search for a potential promoter for gcu14 gene cassette. The 5’UTR from gcu14 were submitted to the promoter predictor programs Neural Network for Promoter Prediction version 2.2 (Berkeley Drosophila Genome Project, http://www.fruitfly.org/index.html) and BPROM (SoftBerry, http://linux1.softberry.com/berry.phtml). Results with the highest scores were selected as candidates for a putative promoter.
The integrons analyzed in this study harbored a rare weak Pc configuration (PcWTGN-10), which presents a C to G mutation 2 bp upstream of the −10 hexamer that causes an abrupt decrease in promoter strength as reported previously7. Considering that transcription initiates from the Pc promoter placed upstream the gcu14-blaGES-1/aacA4 cassette array, both monocistronic and full length polycistronic transcripts could be identified. In fact, Northern blot and hybridization assays revealed a unique signal of approximately 2,300 bases which corresponds to the co-transcription of the entire array (gcu14-blaGES-1/aacA4 ) (Figure 1). This result is in agreement with previous work in which the occurrence of transcripts containing more than one gene cassette was observed by Northern blot analysis1. Moreover, this finding gives support to the lack of attC function in terminating transcription of downstream gene cassettes as demonstrated previously3.

The full length transcript (2,300 bases), corresponding to the entire gene cassette array, hybridized with the GES probe (arrow). The fragment sizes of the RNA marker (Promega) used in RNA electrophoresis are indicated.
This fusion retained both entire coding regions and suffered partial loss of 91 bp at the GES-1 attC site (DQ236170)6. Previous studies demonstrated that the attC region flanked by Left Hand (LH) and Right Hand (RH) domains, which is an imperfect inverted repeat and is missing in the blaGES-1 recombination site, is crucial for cassette mobilization8. Taking into account that the region responsible for stem-loop formation was missing in GES-1 attC and the participation of this site in terminating translation3, our findings indirectly suggested that blaGES-1 and aacA4 translation is occurring in a unique step.
Gene cassettes can be found inserted in integrons or in other secondary sites or free in the cytoplasm as a closed circle, in which the 5’ end (5’ UTR) and the attC recombination site are covalently linked9. As demonstrated previously, several stress conditions could evoke the activation of the SOS response resulting in integron-integrase expression10. Therefore, under stress, the integrase activity increases, favoring the occurrence of integration/excision/rearrangements events. Since the excision event depends on the recognition of the LH and RH domains of the attC site, and that these regions are missing in GES-1 attC, it is expected that the blaGES-1/aacA4 excision occurs only at the aacA4 attC site, and that this structure is excised together as a unique cassette.
Positive results were obtained for the gcu14, gcu14-aacA4 and gcu14-blaGES-1/aacA4 circular forms, but not for blaGES-1 and aacA4 alone, showing that the GES-1 attC is not functional and that the fused gene cassette is excised as a unique cassette. Sequencing assessed the recombination point where excision occurred, confirming the occurrence of free circular forms. The lack of activity of a truncated attC had also been observed before when associated with aadA1011. However, Ramirez and colleagues12 showed that the integrase was able to recognize and mediate excision of a truncated site associated to aadA1, indicating that the genetic context of such truncated sites could influence their role in IntI1 recognition and mobilization.
The relative quantification performed by real time RT-PCR revealed that PS1 and PS26 presented very similar RQ values for gcu14-blaGES-1/aacA4 transcription (Figure 2). This result was expected since integrons from these two strains have the same backbone, including the Pc promoter, and are at the same genetic environment6.
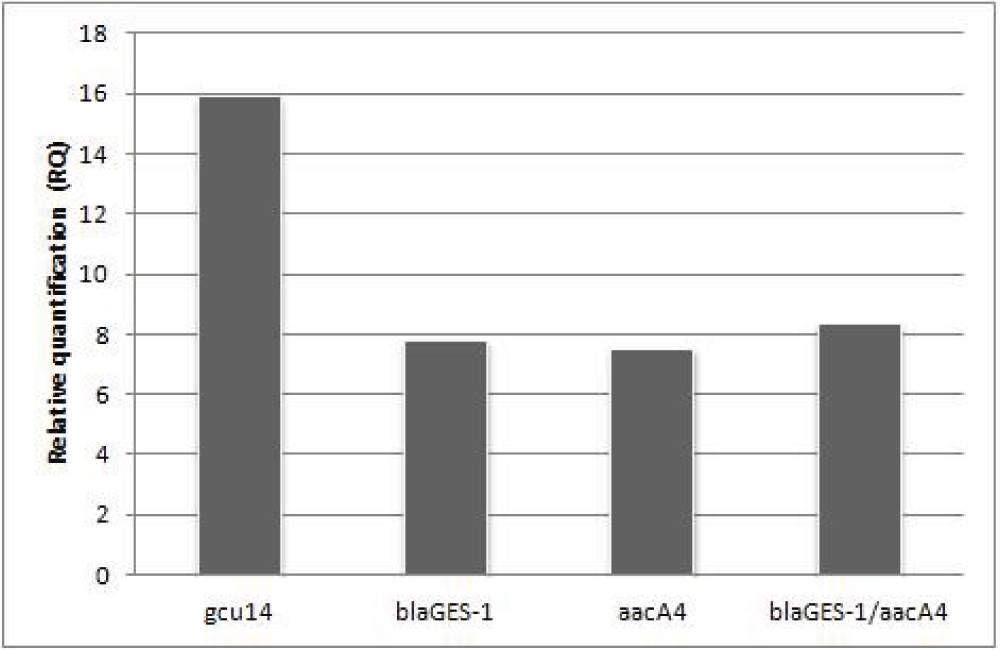
The relative quantification values obtained by real time RT-PCR are indicated for each gene cassette and for the fusion blaGES-1/aacA4 when considered as a unique gene.
gcu14, the first cassette in integrons with the weak PcWTGN-10 configuration, presented approximately two-fold higher transcription when compared to blaGES-1 and aacA4 separately or when the fused cassette blaGES-1/aacA4 was considered (Figure 2). The same RQ value obtained for blaGES-1, aacA4 and the fusion reveals that these two ORFs are transcribed as a unique gene. The lower transcript amount of blaGES-1/aacA4 compared to gcu14 lies on the distance between these gene cassettes and Pc, which is one of the determinants influencing cassette transcription1,10, and it shows the effect of cassette position on expression levels.
A putative promoter for gcu14 (-35 TTGATG [17 bp] -10 TGTTAC) was found 45 bp upstream from its start codon. Therefore, the increased gcu14 transcription could have resulted from a synergistic effect of this putative promoter with Pc. Considering that this putative promoter was found at attI1, which is highly conserved among class 1 integrons, it can be suggested that such a promoter is able to drive transcription of any cassette placed in the first integron position. Moreover, the ORF-11, which enhances the translation efficiency of downstream TIR (translation initiation region)-deficient cassettes inserted in integrons13, was found at the attI1 region preceding the TIR-deficient gcu14 gene cassette. This ORF contained its own Shine-Dalgarno (SD) sequence placed 8 bp upstream of the ATG codon. The ribosome at the ORF-11 stop codon could therefore be carried along the mRNA by lateral diffusion, reinitiating translation at the gcu14 start codon. A potential SD sequence was identified 10 bp upstream of the fused cassette blaGES-1/aacA4. In addition, the loss of the GES-1 attC region, which is involved in stem-loop formation, may enhance the chances of aacA4 translation, since this attC no longer constitutes a physical barrier to ribosome progression3. This deletion also then brings the second gene (aacA4) closer to Pc14. Together, these findings create a scenario for the occurrence of gcu14-blaGES-1/aacA4 expression in PS1 and PS26, which then provides a possible explanation for their resistance profile to β-lactams and aminoglycosides that has been observed elsewhere6.
Fused cassettes have been found in class 1 integrons5,11,12,15–18 ; however, the transcription of such structures has never been addressed. This work showed the transcription pattern of a fused cassette as a polycistronic mRNA and that these unusual structures are excised as a unique cassette.
The sequence of the cassette array composed by the fusion has been deposited in the GenBank database under accession number DQ236170.
ELF and ACPV conceived the study and designed the experiments. ELF carried out the research and prepared the first draft of the manuscript. All authors were involved in the revision of the draft manuscript and have agreed to the final content.
This work was supported by the CNPq and FAPERJ fellowship and Oswaldo Cruz Institute Grant.
We thank those involved in PDTIS platform to enable us to do the sequencing and real-time relative quantification analysis work.








| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 3 (revision) 27 Nov 15 |
read | |
|
Version 2 (revision) 07 Sep 15 |
read | read |
|
Version 1 28 Mar 13 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Besides the comments already made by the reviewers, I would like to specify some points:
- The C to G mutations that converts the usual weak PcW variant into PcWTGN-10 has
... Continue reading Dear Authors,Besides the comments already made by the reviewers, I would like to specify some points:
- The C to G mutations that converts the usual weak PcW variant into PcWTGN-10 has been shown to increase (instead of 'decrease', first part of the Results section).
Response: This sentence was removed.- I have not checked which IntI1 had been used for the excision assay, usually these are made with the most efficient "PcW" IntI1 (IntI1R32_H39); note that in our PLoS Genetics paper (Jove et al. 2010) we evidenced the IntI1 "PcWTGN-10" (IntIP32_H39) were much less efficient for excision of gene cassettes, which could had been taken into account in your discussion.
Response: As properly noticed by you, our Pc configuration is the weak one, and this was changed in the text. However, we agree that including this discussion we will improve our manuscript (lines 182 and 184).- The hypothesis of a synergistic effect between the Pc and P(gcu14) tandem promoters does not fit the observed phenotypes: since the resulting level of transcription observed for the downstream genes is lower I suggest that some transcriptional interferences occur rather than synergistic interactions. By the way in absence of any experimental evidence for the functionality of the gcu14 promoter, such hypothesis is overstated. It would have been interesting to check the level of transcription of the blaGES-1,aacA4 gene cassette when gcu14 is deleted.
Response: we totally agree with this observation, in fact our conclusion is overstated since no experimental assay was performed to test the promoter synergy. Therefore, we remove this part from the text. We also agree that would be interesting to verify the expression of the fused cassette in the absence of gcu14, however it was not the focus of this paper.- First, the class 1 integron sequence of the PS1 strain in Genbank DQ236170 does not display the rare C to G mutation that converts a weak "PcW" promoter into a stronger "PcWTGN-10" as stated in the paper. Consequently you should write "the weak PcW configuration" instead of "weak PcWTGN-10". Also it means that its own IntI1 integrase is likely to be optimally efficient.
Response: Dr. Jové is absolutely right, the DQ236170 accession number presented the PcW configuration and not the PcWTGN-10 as stated in the manuscript, we apologize about this mistake. This was corrected in the text.
- Then, the sequence of the class 1 integron as deposited in the Genbank does not display the complete array of gene cassette (the end of the attCaacA4 is not available) which means that it is ambigous whether there is a downstream third gene cassette or not. Consistently this array of gene cassette, although a novel one, has not been numbered in the INTEGRALL database (http://integrall.bio.ua.pt/?acc=DQ236170)
Response: We know that aacA4 was the last cassette in the array because this variable region was obtained using primers annealing in the attI1 and in the beginning of qacE∆1 from 3’CS. It is true that the aacA4 attC is not completed in the GenBank accession number DQ236170. Therefore, spite of the incomplete attC sequence we are sure that the aacA4 is the last cassette from this array.
- Lastly, as an element of reply to the reviewer Dr Bercot, I would like to notify that the so-called gcu14 GC has not been reported in other reports except in an environmental strain of Citrobacter (and the sequence is not 100% identical, Genbank FM998050).
Response: thanks for this observation. It was included in the text.
Besides the comments already made by the reviewers, I would like to specify some points:
- The C to G mutations that converts the usual weak PcW variant into PcWTGN-10 has been shown to increase (instead of 'decrease', first part of the Results section).
Response: This sentence was removed.- I have not checked which IntI1 had been used for the excision assay, usually these are made with the most efficient "PcW" IntI1 (IntI1R32_H39); note that in our PLoS Genetics paper (Jove et al. 2010) we evidenced the IntI1 "PcWTGN-10" (IntIP32_H39) were much less efficient for excision of gene cassettes, which could had been taken into account in your discussion.
Response: As properly noticed by you, our Pc configuration is the weak one, and this was changed in the text. However, we agree that including this discussion we will improve our manuscript (lines 182 and 184).- The hypothesis of a synergistic effect between the Pc and P(gcu14) tandem promoters does not fit the observed phenotypes: since the resulting level of transcription observed for the downstream genes is lower I suggest that some transcriptional interferences occur rather than synergistic interactions. By the way in absence of any experimental evidence for the functionality of the gcu14 promoter, such hypothesis is overstated. It would have been interesting to check the level of transcription of the blaGES-1,aacA4 gene cassette when gcu14 is deleted.
Response: we totally agree with this observation, in fact our conclusion is overstated since no experimental assay was performed to test the promoter synergy. Therefore, we remove this part from the text. We also agree that would be interesting to verify the expression of the fused cassette in the absence of gcu14, however it was not the focus of this paper.- First, the class 1 integron sequence of the PS1 strain in Genbank DQ236170 does not display the rare C to G mutation that converts a weak "PcW" promoter into a stronger "PcWTGN-10" as stated in the paper. Consequently you should write "the weak PcW configuration" instead of "weak PcWTGN-10". Also it means that its own IntI1 integrase is likely to be optimally efficient.
Response: Dr. Jové is absolutely right, the DQ236170 accession number presented the PcW configuration and not the PcWTGN-10 as stated in the manuscript, we apologize about this mistake. This was corrected in the text.
- Then, the sequence of the class 1 integron as deposited in the Genbank does not display the complete array of gene cassette (the end of the attCaacA4 is not available) which means that it is ambigous whether there is a downstream third gene cassette or not. Consistently this array of gene cassette, although a novel one, has not been numbered in the INTEGRALL database (http://integrall.bio.ua.pt/?acc=DQ236170)
Response: We know that aacA4 was the last cassette in the array because this variable region was obtained using primers annealing in the attI1 and in the beginning of qacE∆1 from 3’CS. It is true that the aacA4 attC is not completed in the GenBank accession number DQ236170. Therefore, spite of the incomplete attC sequence we are sure that the aacA4 is the last cassette from this array.
- Lastly, as an element of reply to the reviewer Dr Bercot, I would like to notify that the so-called gcu14 GC has not been reported in other reports except in an environmental strain of Citrobacter (and the sequence is not 100% identical, Genbank FM998050).
Response: thanks for this observation. It was included in the text.