Keywords
MicroRNAs, PBMC
MicroRNAs, PBMC
The difference between this version and the previous one is in the methods section - cells were stored on ice, prior to extraction and 70% RPMI was used in making the freezing solution.
See the authors' detailed response to the review by Steven O'Reilly
See the authors' detailed response to the review by Stefano Casola
See the authors' detailed response to the review by Kenneth Whitaker Witwer
MicroRNAs (miRNAs) are cell- and therefore tissue-specific, and their expression levels impact protein translation (Sood et al., 2006). Nearly, 2000 microRNA (miRNA) sequences have been identified in humans (Kozomara & Griffiths-Jones, 2014). Numerous studies have reported specific miRNA expression levels in peripheral blood (PB) as markers of disease (Mookherjee & El-Gabalawy, 2013; Patnaik et al., 2012). Although miRNA expression levels could be inevitably influenced by the way that total RNA is extracted, often studies reporting differential miRNA expression levels fail to emphasise the impact of RNA extraction methods. This could at least partially lead to significant controversies and inconsistencies in the literature related to miRNA research.
PAXgene Blood RNA System (PAXgene Blood RNA Tube and PAXgene Blood miRNA Kit [PAXM]) has been the gold standard for PB collection as the stabilising reagent present in the tube prevents RNA degradation and inhibits changes in gene expression due to the collection procedure (Viprey et al., 2012). However, it should be emphasised that the miRNA expression detected in whole blood is the overall outcome from total hematocytes rather than the lymphocyte fraction only. Recent studies have shown that erythrocytes also contain a high proportion of miRNAs (Hamilton, 2010). Bayatti et al. have shown that the presence of globin (globular proteins such as haemoglobin) in PB can impact RNA expression and that globin depletion can decrease total RNA quality and yield in particular when extraction is performed using the PAXgene Blood RNA kit (Bayatti et al., 2014). However, it is necessary to evaluate the miRNA expression levels in an independent cohort, as miRNAs are in general highly stable, and therefore their expression might not be affected by globin treatment. A recent research has reported that miRNA expression levels detected using whole blood correlated to that of PBMCs when using PAXgene Blood RNA System and mirVana miRNA kit (MM), respectively (Mookherjee & El-Gabalawy, 2013). The study did not show that the expression levels are comparable or that they agree with each other using the recommended Bland-Altman method comparison statistical test (Bland & Altman, 1986). This has raised some confusion as the PAXgene Blood RNA system extracts from whole blood while MM from PBMCs. Performing a comprehensive statistical analysis is required when two methods are compared (Burd, 2010).
In this investigation we have measured the expression of miR-146a-5p and miR-155-5p which have extensively been investigated in whole blood and PBMCs due to their critical functions in the innate and adaptive immune system (Curtale et al., 2010; Schulte et al., 2013). We sought to compare miRNA expression levels in peripheral blood collected from 14 healthy volunteers using both extraction methods (PAXM and mirVana PARIS [MP]). We sought to address (i) whether miRNA expression detected in whole blood is comparable to that of isolated PBMCs and (ii) to detect the presence of haemolysis in whole blood and PBMCs. The experimental design for this investigation is depicted in Figure 1.

Peripheral blood was collected into various tubes as illustrated. Total RNA was extracted from whole blood using the PAXgene blood miRNA kit and from PBMCs using the mirVana Paris and mirVana isolation kits, respectively. The total RNA contained the miRNA population which was measured by firstly performing reverse transcription and then quantitative real-time PCR.
Whole blood was collected from 14 healthy volunteers following approval from the Newcastle and North Tyneside 2 Research Ethics Committee (STEMDIAGNOSTICS: REC-07/H0906/131) and informed consent was obtained from every volunteer for both blood collection and miRNA testing. Samples from 14 healthy volunteers (5 males and 9 females) were used to quantify miRNA expression in whole blood and PBMC.
PB (2.5 ml) was collected in PAXgene Blood RNA Tubes (PreAnalytiX GmbH, Switzerland [Catalog No: 762165]) containing an RNA stabilising agent that lysed the blood cells and stabilised the intracellular RNA. The tubes were stored at -20°C until processed. PB was also collected in sodium (Na)-heparin (Sigma, UK) containing tubes for peripheral blood mononuclear cell isolation using graduated centrifugation over Lymphoprep™ (STEMCELL Technologies, Manchester, UK). The cells were then stored on ice before extraction. Isolated PBMCs were cryopreserved by re-suspension in freezing solution containing 70% RPMI 1640 (Sigma-Aldrich, UK), 20% fetal calf serum and 10% dimethyl sulfoxide (NBS Biologicals, UK) and stored in Cryovials at -80°C.
Total RNA was extracted from isolated PBMCs using the (i) mirVana™ PARIS™ kit (MP) and (ii) mirVana™ miRNA Isolation kit (MM) (Ambion, USA [Catalog Nos: AM1556 and AM1560, respectively]) according to the manufacturer’s protocol. PAXgene Blood RNA Tubes were incubated overnight at room temperature to increase the RNA yield. Total RNA extraction from whole blood was performed using the PAXgene Blood miRNA kit (PAXM) (PreAnalytiX GmBh, Switzerland [Catalog No: 763134]), according to the manufacturer’s protocol. Aseptic techniques were followed at all stages of extraction. Total RNA quality and concentration were assessed using NanoDrop ND-1000 spectrophotometer (Thermo Fischer Scientific, MA). The absorbance ratios 260 nm/280 nm and 260 nm/230 nm were analysed to determine the purity of total RNA.
MicroRNA specific cDNA was synthesised from 10 ng total RNA extracted from isolated PBMCs and whole blood using TaqMan MicroRNA reverse transcription (RT) kit (Applied Biosystems, Life Technologies, USA [Catalog No: 4366596]) as per the manufacturer’s protocol. The same RNA concentration was used for all the reactions. Hydrolysis probes were used for cDNA synthesis (Assay IDs: miR-155-5p-5p: 000479, miR-146a-5p: 000468, miR-451-5p: 001141, miR-23a-3p: 000399, SNORD49A [RNU49]: 001005, SNRNP27 [U6]: 001973, SNORA74A [RNU19]: 001003 and SNORD48 [RNU48]: 001006). No-enzyme control (NEC) and a negative template control (NTC) were run for every extraction and RT reaction set. The samples were then run on a thermal cycler at four different holding temperatures: 16°C for 30 minutes, 42°C for 30 minutes, 85°C for 5 minutes and finally at 4°C until storage at -4°C.
Quantitative real-time PCR (qPCR) was performed using the TaqMan method and hydrolysis probes mentioned above (Applied Biosystems by Life Technologies, CA, USA) according to the manufacturer’s protocol. Each sample was run in triplicate and every plate contained the NTC from the RT step and the qPCR step as well as NEC on a 7900HT Fast Real-Time PCR System (Life Technologues, CA, USA).
The qPCR results were analysed using SDS v2.4 software and normalised using SNORD48 as the reference control which was selected by testing a panel of four controls for stable expression within the whole blood and PBMCs. The comparative ∆∆Cq method was used to calculate fold-changes (ΔCq = Cq microRNA of interest - Cq reference control, Relative Quantification [RQ]=2-ΔΔCq and LOG transformed = LOG2RQ). Fold-change was logarithm-transformed as qPCR data are non-linear (exponential), and is transformed to decrease the heterogeneity of variance (McDonald, 2009) and also to identify the outliers present in the data (Rieu & Powers, 2009). Standard curves for three samples from both PAXM and MP were generated and a 95% confidence interval slope of the line was used to calculate the PCR efficiency (E) using the standard formula; E=10-1/Slope and % efficiency = (E-1) × 100. Mean efficiency was then calculated. Results were analysed and plotted using GraphPad PRISM v5.0 software (GraphPad Software, Inc, USA). Mann-Whitney U t-test was used to assess difference between two groups and Kruskal-Wallis one-way analysis of variance (ANOVA) for multiple groups. Spearman’s test was used to determine correlation. Bland-Altman was performed to test whether two methods agreed and if one could be interchanged with another. Significance was set at p<0.05.
Total RNA was extracted using three different extraction methods; PAXM (PAXgene Blood miRNA), MP (mirVana PARIS) and MM (miRVana miRNA) from whole blood and PBMCs. RNA purity was assessed by detecting the absorbance ratios at 260 nm/280 nm and at 260 nm/230 nm. The ratio (260 nm/280 nm) for whole blood was 1.95 – 2.35 and for isolated PBMCs was 2.00 – 2.27 (both MP and MM respectively). The ratios were ≥ 1.8 – 2.0; thus extraction was free from protein contamination which is usually absorbed at 280 nm. Absorbance ratios detected at 260 nm/230 nm showed ratios below the accepted contamination-free range of 1.5 – 2.0 (PBMCs: 0.18 – 1.83 and whole blood: 0.15 – 1.49). Therefore, the samples may have been affected by contaminants absorbed at 230 nm such as guanidine isothiocyanate present in all the three extraction kits. For RNA purity, the peak of each total RNA plot was also analysed as it could indicate contamination by phenol and/guanidine isothiocyanate. PBMCs had plot peaks at 260 nm thus confirming contaminant-free samples. However, peaks at 260 nm were absent for total RNA extracted using the PAXM (Figure 2). This further suggests that guanidine salts may have been the cause of contamination in whole blood samples extracted using PAXM.
NEC and NTC controls were used to ensure that the RT and qPCR reactions were contaminant-free. Each control displayed no amplification (Cq > 36). This was particularly important for total RNA extracted from PBMCs, as they were non-DNase treated. Thus, any amplification in the controls may have suggested either non-specific binding of primers or presence of contamination such as genomic DNA. In addition we calculated the average efficiency of our real-time qPCR reactions (E=97.8%) which confirmed absence of reaction inhibitors such as heparin that was specifically used for the collection of peripheral blood from patients for the downstream PBMC isolation. (See Supplementary Figure 1, that shows there was no contamination in the samples with a Cq value equal to 40 i.e. no amplification). Determining qPCR efficiency is important as inhibitory compounds can affect miRNA expression and result in false positives.
We determined the most appropriate reference gene for this investigation by testing a panel of four controls that have been known for stable expression (SNORD49A [RNU49], SNRNP27 [U6], SNORA74A [RNU19] and SNORD48 [RNU48]) (Dataset a). Our results showed that SNORD48 expression was the most stable in total RNA extracted via both the PAXM (Figure 3A) and MP (Figure 3B) method. Therefore, SNORD48 was used as the reference control to normalise miR-146a-5p and miR-155-5p expression in each sample.
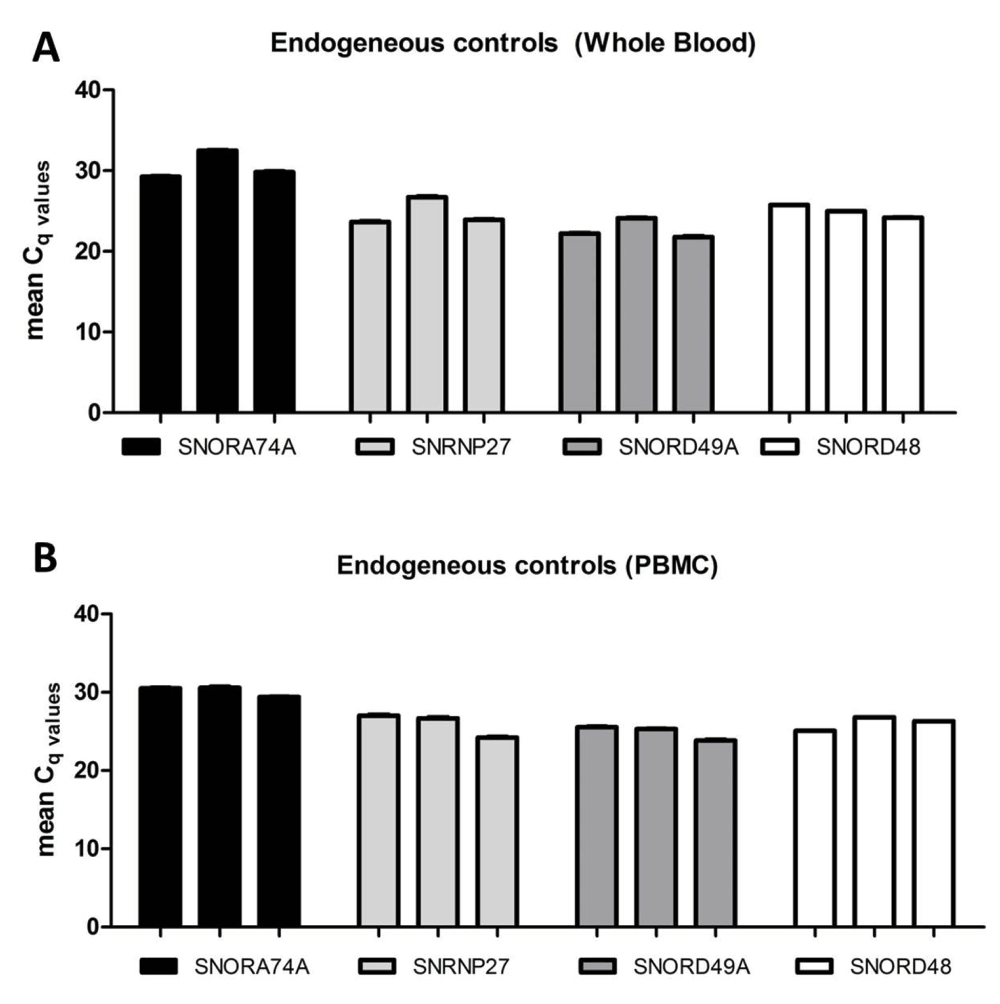
A panel of four stably expressed miRNAs were selected and quantified to identify the most stable control for normalisation of miR-146a-5p and miR-155-5p in paired samples (n=3) (A) whole blood and (B) PBMCs. The standard error of the mean is shown by the error bars for demonstration of technical variability.
Erythrocyte haemolysis has been reported to alter miRNA measurements in whole blood, plasma, serum and tissues (McDonald et al., 2011; Pritchard et al., 2012). The total RNA extracted using the three different methods was examined for degree of haemolysis by quantifying miR-451-5p and miR-23a-3p (Dataset b). Normalised ∆Cq values (miR-23a-3p – miR-451-5p) greater than seven were considered as an indicator of haemolysis. Our results showed that there was a significantly high degree of haemolysis in the total RNA extracted using the PAXM method with the ∆Cq values in the range of 9 – 11. Haemolysis was low in total RNA extracted using either of MP or MM, ∆Cq>3 (Figure 4).

Total RNA extracted using the PAXgene miRNA isolation kit. A threshold of LOG2RQ greater than 7 was indicative of haemolysis (dashed line). The degree of haemolysis was calculated by measuring the difference between miR-451-5p and miR-23a-3p expression. All data (n=5) have been log-transformed. The standard error of the mean is shown by the error bars for demonstration of technical variability.
With inclusion of stringent quality controls, we assessed miR-146a-5p and miR-155-5p expression in total RNA extracted in parallel from whole blood using PAXM (n=14) and PBMCs using MP (n=14) (Dataset c). Our results showed that there was no correlation (Figure 5, miR-146a-5p: r=-0.352, p=0.217 and miR-155-5p: r=0.380, p=0.180) between PAXM and MP in the expression of both miR-146a-5p and miR-155-5p. In a PCR reaction, it is assumed that the target expression doubles at every reaction cycle. Bland-Altman analysis also showed that the two methods did not agree as the bias was greater than 1 which equated to more than one qPCR cycle difference between the two methods (Figure 5A and 5B). Mookherjee et al. had used MM to extract total RNA from PBMCs, then correlated miRNA expression between the PAXM and MM method (Mookherjee & El-Gabalawy, 2013). To eliminate the possibility that using MP was the reason for the non-correlation and disagreement, we tested the three different extraction methods (PAXM, MP and MM) for both miRNAs in a randomly selected cohort of five healthy volunteers (Figure 6) (Dataset d). The results demonstrated that PAXM and MP as well as PAXM and MM did not correlate nor agreed with one another. However, MP and MM methods agreed with each other and could therefore be interchanged as the bias between the two methods for both miR-146a-5p and miR-155-5p was only 0.769 (SD=0.307) and 0.892 (SD=0.802), respectively. Interestingly, normalised miRNA expression was significantly different only between PAXM and MM methods (miR-146a-5p and miR-155-5p: p<0.01). There was higher miRNA expression in PBMCs than in whole blood for both miRNAs (Figure 7).
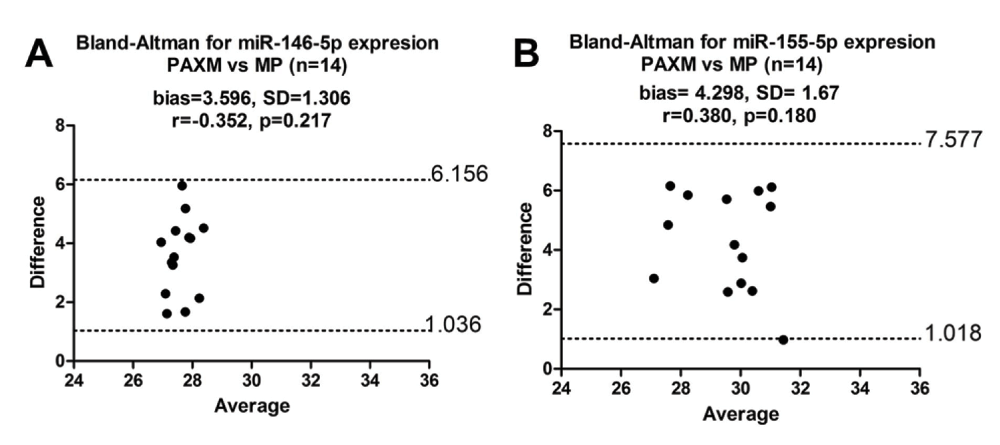
Total RNA was extracted (n=14) using PAXM and MP for (A) miR-146a-5p and (B) miR-155-5p expression. MicroRNA expression is within the limits of agreement but the bias is greater than one showing high disagreement between PAXM and MP. r indicates Spearman correlation. SD: Standard Deviation and bias is the mean difference. Cq values were used for this analysis. Dashed lines show the 95% lower and upper limits of agreement.

The three methods were all compared for miR-146a-5p as (A) PAXM vs MP (B) PAXM vs MM and (C) MP vs MM as well as miR-155-5p (D) PAXM vs MP (E) PAXM vs MM (F) MP vs MM. MicroRNA expression is within the limits of agreement but the bias is greater than one showing high disagreement between PAXM and MP. Bias is lower than one for MP and MM, thus the two methods agree with one another. r indicates Spearman correlation. SD: Standard Deviation and bias is the mean difference. Cq values were used for this analysis. Dashed lines show the 95% lower and upper limits of agreement.

(A) miR-146a-5p and (B) miR-155-5p expression. MicroRNA expression is significantly varied across all the three different groups (p=0.002). MicroRNA expression is higher in PBMCs extracted via either MP or MM method in comparison to whole blood. **p<0.01 and ns: not significant.
MicroRNA expression levels are used to classify diseases and also to distinguish the diseased from the healthy population. However, lack of uniform detection protocols has led to controversies and inconsistencies in miRNA research. There is also lack of recognition for the presence of miRNAs from erythrocytes and other cell-types when using whole blood for total RNA extraction processes and downstream miRNA studies. In most investigations, PBMCs are considered as the major cellular sources for miRNAs. This work was conducted to elucidate the difference between total RNA extracted from whole blood and PBMCs for miRNA expression level studies and also to highlight the importance of protocol standardization.
RT-qPCR was performed to examine whether the expression of miR-146a-5p and miR-155-5p in whole blood and PBMC agreed with one another. Our results showed that there was no agreement between PAXM and both MP and MM for miR-146a-5p and miR-155-5p expression. PBMCs constitute only a fraction of the cells present in PB and therefore lack granulocytes, platelets and erythrocytes (Min et al., 2010). Due to the unique miRNA expression pattern in each cell-type the relative proportions of cells in blood may have an effect on the overall miRNA expression profile and the expression of their protein targets (Min et al., 2010). Some studies have shown that mature miRNA expression signature in erythrocytes is similar to that in whole blood while different when compared to PBMCs (Chen et al., 2008). MiR-451-5p is a marker of erythrocytes (Rasmussen et al., 2010) and miR-23a-3p is unaffected by haemolysis (Blondal et al., 2013). We have shown that there is lower miR-146a-5p and miR-155-5p expression in whole blood compared to PBMCs demonstrating that the total RNA extracted from PBMCs does not reflect that detected in whole blood. Several studies have shown that total RNA yield from whole blood decreases after the use of DNase step in the PAXM protocol (Asare et al., 2008; Bayatti et al., 2014; Debey et al., 2004). However, if PAXM total RNA is not DNase-treated, there may be a possibility of DNA contamination. Contrary to our results, Mookherjee et al. found a linear correlation between miR146a-5p and miR-155-5p expression in whole blood and isolated PBMCs collected from a healthy population (Mookherjee & El-Gabalawy, 2013). In their work, they did not measure the degree of haemolysis in the samples, which may partly explain the discrepancy between the two studies. Furthermore, our study compared both the strength (correlation) and level of agreement between the two methods whilst Mookherjee et al. examined only the correlation (Bland & Altman, 1986). This highlights the importance of performing the correct statistics when two methods are compared with regards to their equivalence and interchangeability (Burd, 2010).
In clinical practice, it is easier to collect PB in PAXgene Blood RNA tubes as they have a shelf-life of two to five years without any RNA degradation. Immediate stabilisation is vital as storage of blood cells induce changes in the miRNA composition (Gaarz et al., 2010). Thus, extraction methods from whole blood must be optimised to either eliminate erythrocyte contamination or consider the expression as cumulative and design downstream experiments for miRNA protein target studies accordingly.
In conclusion, our study showed differences in miR-146a-5p and miR-155-5p expression in isolated PBMCs and whole blood. We suggest that PBMCs are not the ideal source to study and correlate miRNA protein targets where the miRNA expression had been measured in whole blood as the miRNA expression pattern in whole blood is not comparable to that in PBMCs. We also highlight the importance of having a stringent set of technical controls and performing the correct statistics to increase the reliability and reproducibility of miRNA expression studies.
All participants to the study provided informed written consent for molecular testing and publication of the data.
F1000Research: Dataset 1. Data of miRNA extraction methods from whole blood and PBMCs, 10.5256/f1000research.4884.d33496 (Atarod et al., 2014).
S.A performed the experiments, analysed the results and wrote the manuscript. H.S. performed part of the experiments. A.M.D provided constructive comments for the discussion. XN.W interpreted the results and wrote the manuscript. All authors revised the manuscript and agreed to the final content.
This work was supported by the Newcastle University and the FP7 Marie Curie Initial Training Network CELLEurope (Contract No: 315963).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We would like to thank all the volunteers who donated blood for this study. We would also like to thank Dr Clare Lendrem and Dr Kim Pearce for their statistical advice.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 4 (revision) 06 Oct 15 |
|||
|
Version 3 (revision) 22 Jun 15 |
read | read | |
|
Version 2 (revision) 08 Dec 14 |
read | ||
|
Version 1 05 Aug 14 |
read | ||
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)