Keywords
databases, bioinformatics, data integration, molecular biology
This article is included in the Bioinformatics gateway.
databases, bioinformatics, data integration, molecular biology
The use of large-scale data or multi-domain information is becoming a prerequisite in all fields of molecular biology, in light of the advent of high-throughput measurement technologies exemplified by the new generation DNA sequencers, and further driven by the conceptual progress in integrative systems biology approaches. Typical analysis encompasses multiple genes in a pathway or in a regulatory network, uses orthologous gene sets in related organisms, and merges information from multiple-omics layers such as genome, transcriptome, proteome, and metabolome (Arakawa & Tomita, 2013). Bioinformatics researchers therefore need to collect and integrate data from a variety of sources, each with diverse syntax, semantics, protocols, identifiers and naming conventions (Bhagat et al., 2010; Brazas et al., 2012; Katayama et al., 2010). This data integration issue is one of the key challenges in the field of bioinformatics (Stein, 2002; Stein, 2008). While the integration of web services under standardized protocols has seen a sound progress over the last few years (Katayama et al., 2011), data integration with efficient cross-domain queries still requires the use of large-scale data warehouses such as BioMart (Smedley et al., 2009) and InterMine (Smith et al., 2012).
Since the majority of biological databases are well curated with cross-references, related information can be retrieved ad hoc from dispersed databases using hyperlinks. In order to facilitate such processes, web services that collect and provide the cross-reference information from different databases (Diehn et al., 2003; Wu et al., 2013) as well as ID conversion services that assist cross-referencing have been developed (Cote et al., 2007). MyGene.info, for example, provides rapid programmatic access through a RESTful interface for gene-centric queries to retrieve cross-reference information from numerous databases. Gene-centric aggregation, which integrates databases using genes as the primary key, is a highly efficient approach in molecular biology, since the majority of databases have some sort of connection to genes or proteins, due to the success of the “central dogma” of molecular biology. Ideally, a database should be free from predefined schema or primary keys, and should have controlled syntax and semantics. Semantic Web initiatives are therefore collaboratively aiming to provide such framework through HyperText Transfer Protocol (HTTP) with Resource Description Framework (RDF) and Web Ontology Language (OWL) (Katayama et al., 2013). At the current state of Semantic Web technologies, however, cross-domain queries require extensive reasoning or manual curation of ontologies, and the cross-reference-based approach still has an advantage in terms of user experience with lower latency.
Cross-reference services usually provide database name and identifiers that do not explicitly define the actual location of the data. Moreover, gene-centric data aggregation services usually do not allow querying of gene sets. To this end, here we describe a new RESTful service named G-Links, which gathers Uniform Resource Identifiers (URI) from more than 100 databases in a gene-centric manner, and provide querying interface based on gene sets for hundreds of species. G-Links can be used programmatically as text data, from Semantic Web services, or from graphical HTML pages.
G-Links is implemented with Perl programming language and MySQL 5.0, and has a straight-forward RESTful user interface. It internally resolves cross-references in four steps: ID conversion, retrieval of cross-references, filtering and extraction, and formatting of output. G-Links stores all cross-reference information in a gene-centric manner, and for this purpose, it utilizes UniProt IDs as the primary key. Therefore, G-Links first converts the user input to UniProt ID by ID conversion, based on 80 databases supported by UniProt ID Mapping Service (Huang et al., 2011). When a nucleotide or amino acid sequence is given as the query, G-Links searches the corresponding UniProt IDs by sequence similarity search using BLAT (Kent, 2002) against Swiss-Prot database (Bairoch et al., 2004), and when RefSeq ID for bacterial genomes or taxonomy ID is used as the input, G-Links collects all UniProt IDs of genes within the given species based on UniProt taxonomy (http://www.uniprot.org/taxonomy/). In the second step, G-Links collects all text annotations and database cross-references about the gene of interest, gathered from over 130 databases. Here the mapping to Gene Ontology slim (Harris et al., 2004) is pre-computed using map2slim (http://search.cpan.org/~cmungall/go-perl/scripts/map2slim), and resulting URLs for over 30 RESTful bioinformatics analysis web services supported by the G-language Web Services (Arakawa et al., 2010) and Keio Bioinformatics Web Service (KBWS) (Oshita et al., 2011) are generated on-the-fly. KBWS is an European Molecular Biology Open Software Suite (EMBOSS) (Rice et al., 2000) associated software package for accessing popular bioinformatics web services such as BLAST. All cross-references include the URI of the actual location of data, often expressed as Persistent Uniform Resource Locators (PURLs). Retrieved gene set and annotations are optionally filtered in the third step according to user input, and are formatted in the specified output format in the last step.
G-Links is available at http://link.g-language.org/ as a RESTful web service, which is suited for resource-centric access and highly accessible via HTTP. Users can rapidly retrieve annotations and cross-references related to a given gene ID, taxonomy ID, or raw sequence data by simply accessing a certain URL. An overview of the URL syntax is presented in Figure 1. For example, the URL to retrieve all annotations and cross references related to the human BRCA1 gene (UniProt ID: BRCA1_HUMAN) is simply http://link.g-language.org/BRCA1_HUMAN (Figure 2). The ID of gene used in this query can be any of the identifiers used in 80 databases supported by G-Links. Programmatic access to this URL can retrieve all 653 annotations and cross-references within 0.2 seconds (tested on dual Xeon X5470 server). G-Links automatically adjusts the output format according to the user context, and outputs the results in human-readable interactive HTML format when accessed from modern HTML browsers, or in Tabular Separated Values (TSV) text format for programmatic access. The HTML format displays a gallery of image resources on the top, such as the pathway maps from KEGG database (Kanehisa et al., 2012), co-expressed gene network from COXPRESdb (Obayashi et al., 2013), and protein 3D structure from Protein Data Bank (Rose et al., 2013), followed by a long table of text annotations and cross-references including database name, ID, and resource URL. Table 1 shows an overview of the categories of databases and web services supported by G-Links output (see http://link.g-language.org/input_list and http://link.g-language.org/output_list for complete listings). In addition to the human-friendly HTML format and computer readable TSV as well as JavaScript Object Notation (JSON) output, G-Links supports RDF/XML and Notation3 (http://www.w3.org/TeamSubmission/n3/) formats, so that the query results can be readily integrated with Semantic Web technologies. For RDF and Notation3 predicate information is given by EMBRACE Data and Methods (EDAM) ontology.
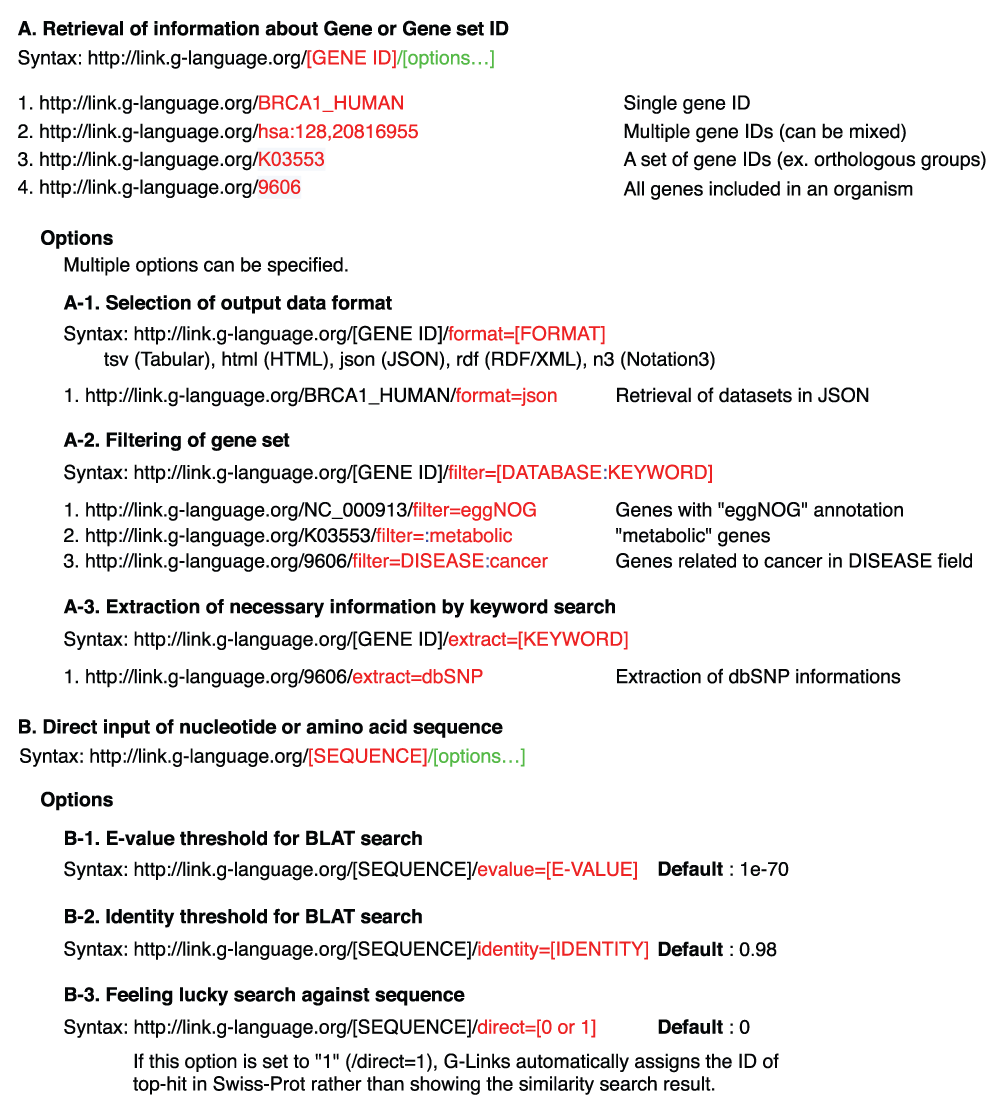
G-Links is implemented as a RESTful service that can be queried by altering the URL. Full documentation and example queries are available at http://www.g-language.org/wiki/glinks.
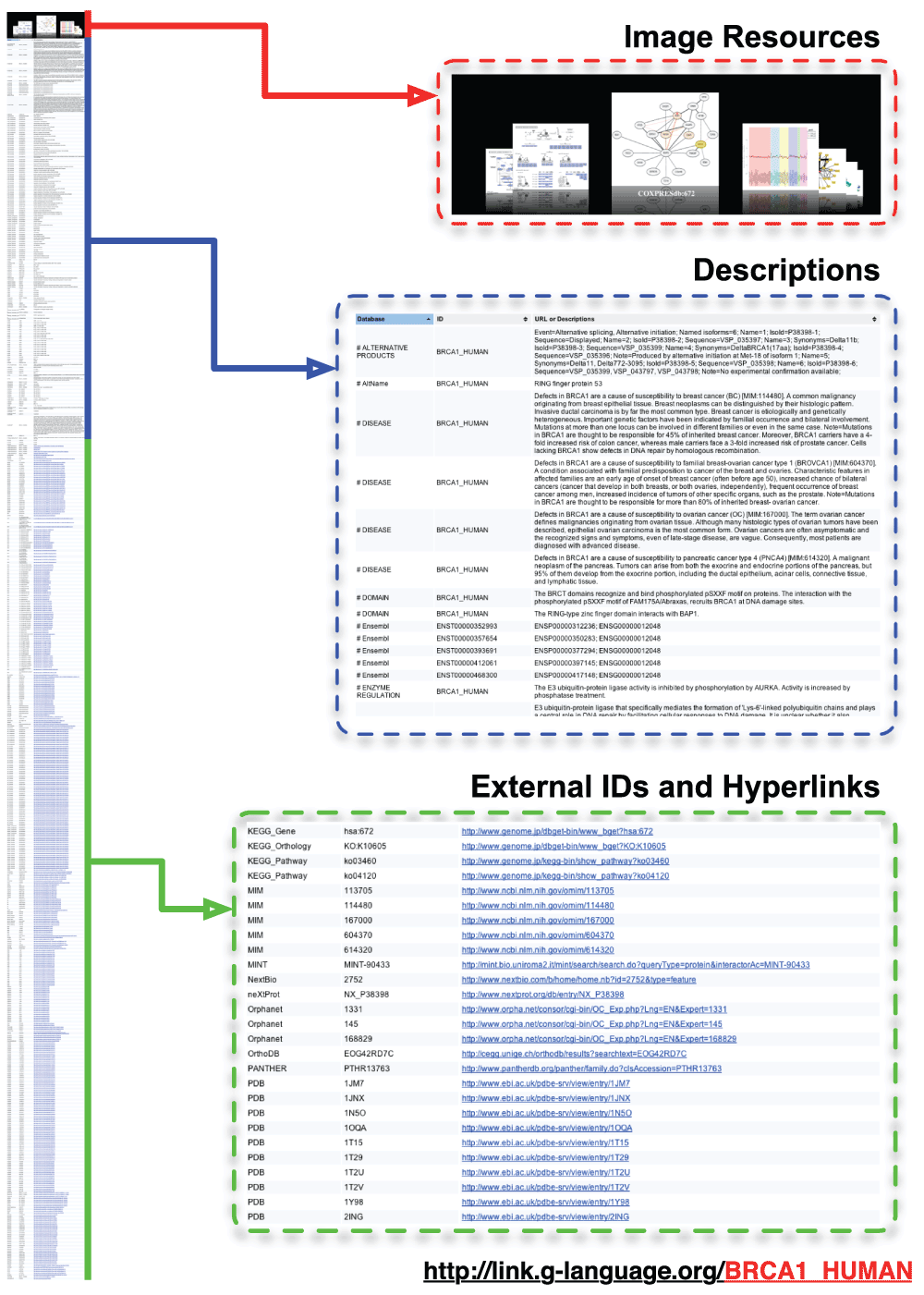
By default, access to G-Links with web browsers displays the results in interactive HTML, with related image gallery implemented with CoverFlow (http://imageflow.finnrudolph.de/) on the top, followed by a large table of annotations and cross-references.
Detailed list of Input/Output databases are available at http://link.g-language.org/input_list and http://link.g-language.org/output_list.
One of the advantages of G-Links over existing cross-referencing services is its ability to retrieve information related to gene sets or all genes of organisms, and to filter out non-related genes by keyword search (filter option) or to extract necessary fields (extract option). Using the filtering option, users can retrieve only the subset of genes related to the given keyword. For example, retrieval of all human (taxonomy ID: 9606) genes having GO slim function including the word “transport” is as simple as http://link.g-language.org/9606/filter=GOslim_function:transport/format=tsv/. Similarly, extraction of only the “DISEASE” annotation of BRCA1 gene is simply http://link.g-language.org/BRCA1_HUMAN/extract=DISEASE. Multiple filtering and extraction conditions can be specified using “|” (vertical bar) as the separator, in order to formulate complex queries. For example, retrieval of SNP information from dbSNP and SNPedia for human genes with known polymorphisms related to breast and ovarian cancer in tabular format is queried as http://link.g-language.org/9606/format=tsv/filter=DISEASE:cancer/filter=:breast|:ovarian|:snps|:polymorphisms/extract=dbSNP|SNPedia.
By serving as a data hub of linked open biological data, G-Links can be a starting point in retrieval of gene-centric information. Users can quickly obtain related links and annotations of a gene of interest either graphically via HTML or programmatically via REST interface, such as the orthologs, Gene Ontology terms, protein structure, pathways, SNPs, and publications.
G-Links is a RESTful service with base URL http://link.g-language.org/. Detailed documentation is available at http://www.g-language.org/wiki/glinks including service description, syntax, list of all available options, example queries (URLs) and sample scripts for programmatic access in Perl, Ruby, Python, and Java. Comprehensive lists of supported input/output databases and web services are available at http://link.g-language.org/input_list and http://link.g-language.org/output_list. Internal database of G-Links is regularly updated every month, and the source code is freely available from GitHub repository (http://github.com/cory-ko/G-Links).
http://dx.doi.org/10.5072/zenodo.12701 (Oshita & Arakawa, 2014)
KO and KA conceived and designed the software, and KO implemented the software. MT provided supervision for the study. KO and KA drafted the manuscript, and all authors were involved in the revision of the draft manuscript and have agreed to the final content.
This work was supported by KAKENHI Grant Number 222681029 from the Japan Society for the Promotion of Science (JSPS), and by funds from the Yamagata Prefectural Government and Tsuruoka City.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The development of web services described in this work was significantly facilitated by the DBCLS BioHackathon 2012 hosted by the National Bioscience Database Center/Database Center for Life Science (NBDC/DBCLS). We thank the members of G-language Project at the Institute for Advanced Biosciences, Keio University, and elsewhere, for their extremely valuable feedback and support.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 18 Nov 15 |
read | |
|
Version 1 19 Nov 14 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)