Introduction
Numerous web applications exist for visualisation of biological data. Data can be prepared for visualisation using a variety of formats, one of which is the widely used wiggle (wig) file. A wiggle file contains text that defines either a feature or a data track. The wiggle format was developed by the UCSC genome browser1 and then quickly adopted by other initiatives2,3. Web applications such as genome browsers rely heavily on JavaScript, a popular language for processing and rendering client-side information in a web browser. Despite their widespread use in bioinformatics, biological web applications are usually implemented with no standard reutilisation guidelines in mind, hence BioJS was developed4.
BioJS is an open source JavaScript library of components for the visualisation of biological data on the web. Here we present wigExplorer, a standard, portable BioJS component designed to easily render wig data format files. wigExplorer can be integrated and controlled from other applications. To our knowledge, this is the first modular component to visualise wig data that complies with BioJS standards.
The wigExplorer component
wigExplorer is fully integrated in the BioJS project. It follows the standards set by the BioJS registry5, a centralised repository of BioJS components hosted at the European Bioinformatics Institute (EBI). Having wigExplorer in the BioJS registry is advantageous because it promotes i) easy discoverability for the component, ii) collaborative development with other members of the BioJS community and iii) reutilisation by third party applications. In the BioJS registry, component APIs are exposed, i.e., events and methods are defined and documented so that other BioJS components can interact with each other. By following these conventions, wigExplorer is able to interact with other components on the same web page, enriching the overall experience for the user. The code below shows how to incorporate wigExplorer into a web application. Only three configuration elements are needed: the target HTML element in which the component will be rendered, the background colour of the component, and the file path containing the wig data.
var instance = new Biojs.wigExplorer({
target: "YourOwnDivId",
selectionBackgroundColor: ʼ<background-colour>ʼ,
dataSet: "<path–to–file>"
});
wigExplorer uses D3.js, the data-driven documents JavaScript library6, to generate graphical representations from wig data. D3.js handles the manipulation of the data documents, the reading of wig data as text format and their conversion to an area chart format. To control the visual aspect of the wig data, wigExplorer contains simple controls for zooming and panning.
Application
wigExplorer can be used to visualise genomic data in different ways. An application is shown in Figure 1, depicting single nucleotide polymorphism (SNP) density data from a genomic annotation in the tomato genome. Here chromosome 2 is zoomed in to show the genomic interval contained between position 3M and 47M. The SNP density data contained in the wig data file are presented as bins, where the Y axis indicates the number of SNPs contained in each bin. The screenshot shows a dramatic change in the density of SNPs at the 24M bin mark of the chromosome, suggesting a potential boundary for an introgression segment introduced from a closely related tomato species. Other potential applications of wigExplorer may involve the visualisation of gene expression and alignment data.
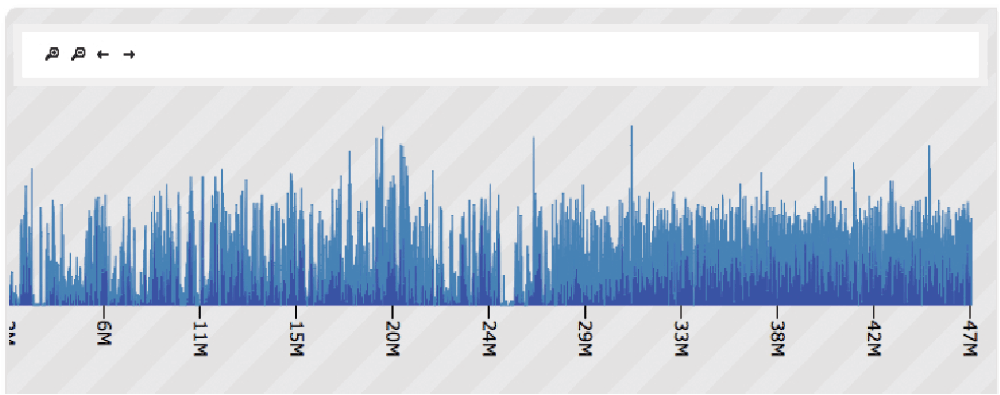
Figure 1.
wigExplorer view of tomato variety Heinz chromosome 2.
The top controls are designed to scroll sideways. Peaks show SNP density of 1kb size bins. A change of SNP density can be observed around the 29M mark, with a slightly greater density of SNPs on the right, indicative of a potential introgression segment from another related species.
We are aware that third party browsers are also using wigExplorer. A screenshot of the TGAC Browser7 is shown in Figure 2 using wigExplorer to depict Myzus spp. scaffold 1 zoomed in between regions 714K and 727K. Here strand-specific RNA-Seq paired-end read coverage is shown as a wig track. The track below shows a closely related annotated species gene set for comparison. This comparison suggests a potential gene extension in both forward and reverse orientation.

Figure 2. An example of wigExplorer integration using the TGAC Browser.
The wigExplorer track shows read coverage in Myzus spp. for scaffold 1. Forward and backward strands are depicted in red and blue respectively. Evidence genes from a closely related species are displayed in the track below.
Conclusions
The wigExplorer component provides a platform to visualise biological data in wig format. wigExplorer can be easily integrated with other web components or extended to provide new functionality. We expect this component to be particularly useful for visualisation in a variety of data types such as SNP density, alignments and gene expression. Like any other BioJS component, wigExplorer requires little technical knowledge for its utilisation.
Software availability
Zenodo: wigExplorer, a BioJS component to visualise wig data, doi: 10.5281/zenodo.77218
GitHub: BioJS, http://github.com/biojs/biojs;
Author contributions
AT and RJ developed the code for wigExplorer. MC and GK created the user cases for Figure 1 and Figure 2 respectively. AT, RD, MC and GK wrote the paper.
Competing interests
No competing interests were disclosed.
Grant information
AT, GK, MC and RD were supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC) National Capability Grant (BB/J010375/1) at TGAC.
Acknowledgements
We are grateful to all BioJS developers who have contributed their work under an open source license. We are grateful to David Swarbreck at TGAC for his advice on the data shown in Figure 2.
Faculty Opinions recommendedReferences
- 1.
Kent WJ, Sugnet CW, Furey TS, et al.:
The human genome browser at UCSC.
Genome Res.
2002; 12(6): 996–1006. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 2.
Donlin MJ:
Using the Generic Genome Browser (GBrowse).
Curr Protoc Bioinformatics.
John Wiley and Sons, Inc., 2009. PubMed Abstract
| Publisher Full Text
- 3.
Skinner ME, Uzilov AV, Stein LD, et al.:
JBrowse: A next-generation genome browser.
Genome Res.
2009; 19(9): 1630–1638. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 4.
Gómez J, García LJ, Salazar GA, et al.:
BioJS: an open source JavaScript framework for biological data visualization.
Bioinformatics.
2013; 29(8): 1103–1104. PubMed Abstract
| Publisher Full Text
| Free Full Text
- 5.
BioJS: registry. http://www.ebi.ac.uk/Tools/biojs/registry/, 2013. Reference Source
- 6.
D3.js data-driven documents. http://d3js.org, 2012. Reference Source
- 7.
Thanki AS, Bian X, Davey RP, et al.:
TGAC Browser: visualisation solutions for big data in the genomic era. 2013. Reference Source
- 8.
Thanki AS, Jiminez RC, Kaithakottil GK, et al.:
wigExplorer, a BioJS component to visualise wig data.
Zenodo.
2014. Data Source


Comments on this article Comments (0)