Keywords
DNA, mice, solutions, eukaryotic cells
DNA, mice, solutions, eukaryotic cells
Infections by Shiga toxin-producing Escherichia coli (STEC) strains are a serious public health concern, resulting in diarrhea, hemorrhagic colitis, and haemolytic uremic syndrome (HUS).
Stx is the main virulence factor in STEC strains. The stx gene is present in the genome of prophages, which are similar to the bacteriophage lambda found in the lysogenic form of various E. coli strains. Previously we reported that the native promoter of the Stx-encoding gene can drive expression of the toxin in eukaryotic cells in both in vivo and in vitro conditions1,2.
Many questions remain unanswered with regard to the mechanism by which STEC infection causes HUS. In particular, we are interested in understanding how Stx enters the systemic circulation and why only very small numbers of bacteria are sufficient to induce HUS in humans.
Based on our previous observations that the native stx gene promoter is active in host cells, we seek to understand the role that bacteriophages play in the pathogenesis of STEC strains. Recently, it was reported that bacteriophages carrying the stx gene are required for the development of HUS in the murine model3. We hypothesise that eukaryotic host cells might be transduced with and/or infected by Stx-encoding bacteriophages, leading to Stx dissemination in vivo to enter the systemic circulation. This would also explain why very small numbers of bacteria are sufficient to develop HUS.
In order to test whether bacteriophages are responsible for the induction of HUS, we used an anti-bacteriophage agent to inactivate them. Chitosan, a linear polysaccharide polymer obtained after the deacetylation of chitin, the structural element in the exoskeleton of crustaceans, possesses strong antimicrobial activity against several pathogenic microorganisms4. Its antiviral activity was reported on the bacteriophage c2, which infects Lactococcus strains, and on bacteriophage MS2, which infects E. coli5 without affecting significantly the growth of the bacterial culture6. In order to test our hypothesis, which would make Stx-encoding bacteriophages a new target for preventing and treating STEC infections, we used chitosan as an anti-bacteriophage agent in vitro and in vivo.
Inactivation of bacteriophages was observed in vitro after incubation with chitosan, inhibiting both the infection of, and replication in bacterial cells, and the transduction of eukaryotic cells.
GFP dissemination was significantly reduced in mice treated with chitosan following infection with a non-pathogenic strain carrying a bacteriophage in which the stx gene was replaced by the GFP-encoding sequence. Last, preliminary results showed partial protection by chitosan in vivo of mice infected with STEC.
These results contribute to understanding STEC infections, posing implications for a similar scenario to occur in other infections caused by bacteria carrying lysogenic bacteriophages.
C600ΔTOX:GFP, a lysogenized C600 strain carrying the 933W bacteriophage in which the stx gene was replaced by the gfp sequence, was generously provided by Dr. Alison Weiss7. EDL933W, an enterohemorrhagic E. coli (EHEC) strain carrying the wild-type bacteriophage from which C600ΔTOX:GFP was obtained, was generously provided by Dr. Luis Carlos de Souza Ferreira, LDV-USP, Brazil.
Baby BHK-21 cells (Syrian hamster kidney fibroblasts from the American Type Culture Collection) cells were grown on 12-well plates (Nunc) in complete medium (10% fetal bovine serum in DMEM medium, Gibco, USA) for use in the transduction assay.
C600ΔTOX:GFP was generously provided by Dr Alison Weiss7. This is a non-pathogenic phage resulting from purified 933W bacteriophage in which stx gene was replaced by gfp sequence (ϕ ΔTOX:GFP). Phages at a multiplicity of infection (M.O.I) equal to 1 were added to BHK-21 cells cultured the day before on 12 wells plate (Nunc). BHK-21 cells were counted with a Neubauer camera, and bacteriophage titer was measured by the titration assay as described below. Transduction of BHK-21 cells was enhanced by centrifugation at 1000 × g for 10 minutes at room temperature as previously reported1. After incubation at 37°C for 3 hours, the phage-containing medium was removed. Cells were washed twice with phosphate buffered saline (PBS) and then incubated in complete DMEM medium (Gibco, USA). Twenty four hours post-transduction, cells were washed with PBS, harvested and centrifuged at 2655 × g for 15 minutes. DNA was harvested from pellets after incubation for 5 minutes at 98°C in lysis solution (Tris pH8 50mM, SDS 2%, Triton-X100 5%) and the harvested DNA was used for PCR. Primers: Up-R 5′CCGCTCGAGACTAGTGCAAAAGCGAGCCTGGTAAATAAATATG3′; Up-D 5′GGAATTCCATATGCTCGTTGAGGCATATGAAAATCAGAC3′. The reaction was run in a Eppendorf Termocycler at an initial 92°C for 120 seconds and then at 92°C for 20 seconds and 60°C for 20 seconds and 72°C for 120 seconds for 35 cycles using primers giving a fragment of 1310 bp on the upstream region of gfp gene into the bacteriophage genome.
The C600ΔTOX:GFP strain was grown in Luria Broth (LB) plus 10 mM CaCl2 and chloramphenicol (Sigma) (15 μg/ml final concentration) overnight (ON) at 37°C under agitation. The ON culture was diluted to OD600nm = 0.1 in LB plus 10 mM CaCl2 and chloramphenicol (Sigma) (15 μg/ml final concentration). Induction was carried out by adding ciprofloxacin to a final concentration of 40 ng/ml8. Bacteria were incubated for 6 hours at 37°C under agitation. Cultures were then centrifuged at 5000 rpm for 15 minutes. The bacteriophage-containing supernatant was filtered with 0.2 μm filters and kept at 4°C until the titration assay was performed.
E. coli strain Y1090 (ATCC 37197) was grown in LB plus ampicillin ON at 37°C under agitation. The ON culture was diluted 1:100 in LB plus ampicillin and incubated for 2 hours at 37°C under agitation. At the end of the incubation, 500 μl samples of E. coli Y1090 were incubated with 5, 50 and 100 μl of a suspension containing bacteriophages for 30 minutes at room temperature. At the end of this incubation, 3 ml of Top Agar (Tryptone 1%; NaCl 0.5%; Agar 0.7%) plus CaCl2 (10 mM final concentration) was added, and plated on LB-Amp agar plates. Plates were incubated at 37°C and lysis plaques were visually counted.
ϕΔTOX:GFP was incubated with 5 mg/ml of a chitosan (Sigma 448877) solution in phosphate buffer 10 mM, at pH = 7 for 10 minutes at room temperature, and bacteriophage titers were measured as described in titration assay section.
Chitosan was also used in the bacteriophage induction assay described above. Chitosan was added 2 and 4 hours post-induction and bacteriophage titers were analyzed at 6 hours post-induction.
BALB/c mice were bred in-house at the animal facility of the Microbiology Department of the São Paulo University, Brazil. The experimental protocol of this study followed the ethical principles for animal experimentation adopted by the Brazilian College of Animal Experimentation (COBEA) and was approved by the Ethics Committee on Animal Experiments of the Institute of Biomedical Sciences (Protocol number 106), University of São Paulo, in accordance with the principles set forth in the Guide for the Care and Use of Laboratory Animals (National Institutes of Health, 1985).
Male mice aged 6 weeks (18 to 20 g) were used for the In Vivo Imaging System (IVIS). Immature male and female DBA-2 mice (17–21 days of age, approximately 8–11 g body weight) were used immediately after weaning for the infection assays with EDL933W strain (n = 4). Mice were maintained under a 12-h light-dark cycle at 22 ± 2°C and fed a standard diet and water ad libitum.
Immature male and female DBA-2 mice (17–21 days of age, approximately 8–11 g body weight) were used immediately after weaning for the infection assays (n = 4).
E. coli EDL933W (ATCC 43895) was used for infection of mice following the protocol previously reported by Brando and collaborators8. Briefly, E. coli EDL933W was grown in Tryptic Soy Broth (TSB, DIFCO, BD) ON at 37°C. The ON culture was centrifuged at 14000 rpm for 15 minutes and the bacterial pellets were washed twice in PBS. Pellets were resuspended to have a final concentration of 3 × 1012 CFU/100 μl per mouse.
The bacterial suspension was delivered directly into the stomach of mice after 8 hours of food starvation, via a 5-French paediatric feeding tube. After 4 hours of ingesting the bacterial suspension, mice were given food and water. Control animals received 100 μl of sterile PBS. Survival was observed for one week. Both groups were composed by 4 animals.
To analyze the effect of chitosan in vivo, immature male and female DBA-2 were infected as described previously and treated with 100 μl of a chitosan solution at a concentration of 5 mg/ml, orally administered 2 hours after infection. Survival was observed for one week.
Two-month old BALB/c mice were used to infect orally with C600ϕΔTOX:GFP. Bacteriophage induction in vivo was performed with ciprofloxacin as described immediately below. After 2 hours of induction with ciprofloxacin, 100 μl of chitosan solution at a concentration of 5 mg/ml was administered orally to the mice and GFP dissemination by IVIS was analyzed.
This time, we used two-month old BALB/c mice. An ON culture of C600:ϕΔTOX-GFP was used to infect them. The ON culture was centrifuged at 14000 rpm for 15 minutes at 4°C. The pellet was washed with PBS and centrifuged again at 14000 rpm for 15 minutes at 4°C. The pellet was resuspended in a solution of 20% sucrose to have a concentration of 1 × 109 CFU/mouse. Mice were inoculated orally with strain C600:ϕΔTOX-GFP and in vivo bacteriophage excision was induced following the procedures described by Zhang and collaborators8. The mice were sacrificed with CO2 inhalation 24 hours after bacterial inoculation. Blood, spleens, kidneys, lungs, brains, intestines, hearts and livers were harvested by surgical removal and kept in PBS solution and evaluated for GFP expression using the IVIS system. To determine the effects of chitosan in vivo, the mice received 100 μl of a chitosan solution at a concentration of 5 mg/ml.
Statistical significance between treatments and controls was analyzed using the Prism 5.0 software (GraphPad Software), and the P value is indicated by asterisks in the figures.
All other data correspond to the means ± standard errors of the means (SEM) for individual mice. Statistical differences were determined using the one-way analysis of variance (ANOVA).
Bacteriophage lytic induction was triggered in E. coli C600ΔTOX:GFP using ciprofloxacin8. We observed a significant decrease in the optical density of the bacterial culture after addition of the antibiotic and the release of phages into the culture supernatant (Figure 1, panel A and B). The bacteriophage titer was analyzed at different time points and a significant increase was observed after induction (Figure 1, panel B). The effect of chitosan as an anti-bacteriophage agent was also examined. To this aim, we added chitosan at a final concentration of 5 mg/ml to the bacterial culture 2 or 4 hours post-induction, and we observed the complete inactivation of the ϕΔTOX:GFP, without measurable toxic effects to the bacterial strain (Figure 1, panels A and B).
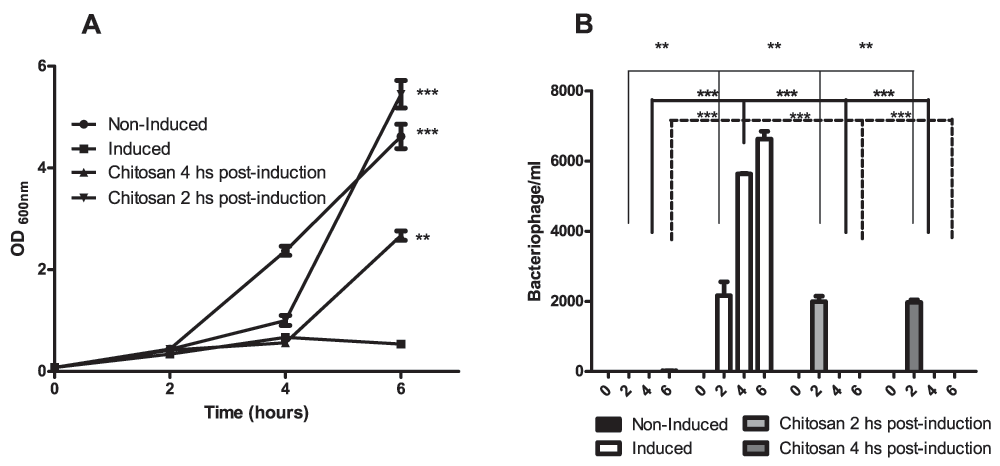
A. Growth curve: C600ΔTOX:GFP was induced with ciprofloxacin and the optical density was measured at 600 nm at 0, 2, 4 and 6 hours after induction. Non-induced C600ΔTOX:GFP was used as control. Chitosan was added at 2 or 4 hours after induction. B. Bacteriophage ϕΔTOX:GFP titer: bacteriophage titers were analyzed at 0, 2, 4 and 6 hours post-induction. Chitosan was added at 2 or 4 h post-induction. Asterisks represent P<0.05.
We previously reported the capacity of ϕΔTOX:GFP to transduce macrophages in vitro1. To further evaluate the ability of chitosan to inhibit bacteriophage transduction of mammalian cells, BHK cells were transduced for 3 hours with ϕΔTOX:GFP, ϕΔTOX:GFP plus chitosan or ϕΔTOX:GFP treated with DNAse. Addition of DNAse to the bacteriophage sample would preclude any free DNA in the bacterial lysates prior to the transduction of cells. Untreated cells were used as a control. As shown in Figure 2, the bacteriophage DNA was detected by PCR in mammalian cells, showing the capacity of the virus to transduce this cell line. However, when BHK cells were transduced with bacteriophages pre-incubated with chitosan, no phage DNA was detected, confirming the inactivating action of chitosan on bacteriophages. Bacteriophage DNA was also detected in cells transduced with ϕΔTOX:GFP treated with DNAse (Figure 2).
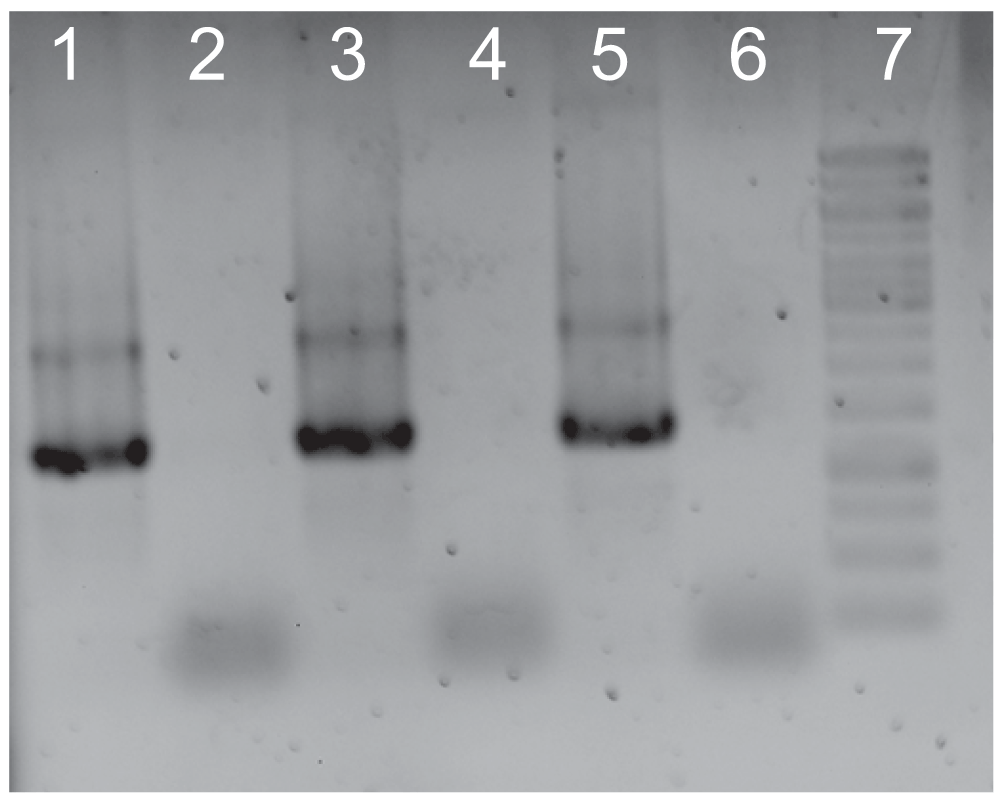
A. PCR on DNA extracted from eukaryotic cells: 24 hours after transduction, BHK cells were washed and treated with Trypsin-EDTA solution. DNA was extracted and PCR was performed. Line 1: Cells transduced with ϕΔTOX:GFP. Line 2: Cells transduced with ϕΔTOX:GFP plus chitosan. Line 3: Cells transduced with ϕΔTOX:GFP previously treated with DNAse. Line 4. Untreated cells. Line 5. Positive control (ϕΔTOX:GFP DNA). Line 6. Negative control. Line 7. 1 kb ladder (Invitrogen).
To demonstrate the in vivo behavior of bacteriophages, mice were infected with the lysogenic E. coli C600ΔTOX:GFP strain, followed by oral administration of ciprofloxacin 1 hour or 2 hours later. In order to evaluate the effect of chitosan in vivo, a group of mice was administered with chitosan 2 hours post-induction and a control group of uninfected mice was evaluated for auto-fluorescence background control in each organ. Twenty four hours after infection, organs were harvested and examined using the IVIS. As shown in Figure 3, GFP was detected in the intestine, liver and, to a lesser extent, kidney of mice orally infected and treated with ciprofloxacin. Remarkably, the addition of chitosan 2 hours after infection caused a sharp decrease in GFP detection in organs of mice orally infected with the E. coli strain C600ΔTOX:GFP (Figure 3, panels A and B), indirectly indicating reduction of bacteriophages in the cells, GFP release and dissemination. Moreover, viable phages were detected via the lysis plaque assay in intestine homogenates and blood samples of infected mice, in which bacteriophages were induced by ciprofloxacin (data not shown).
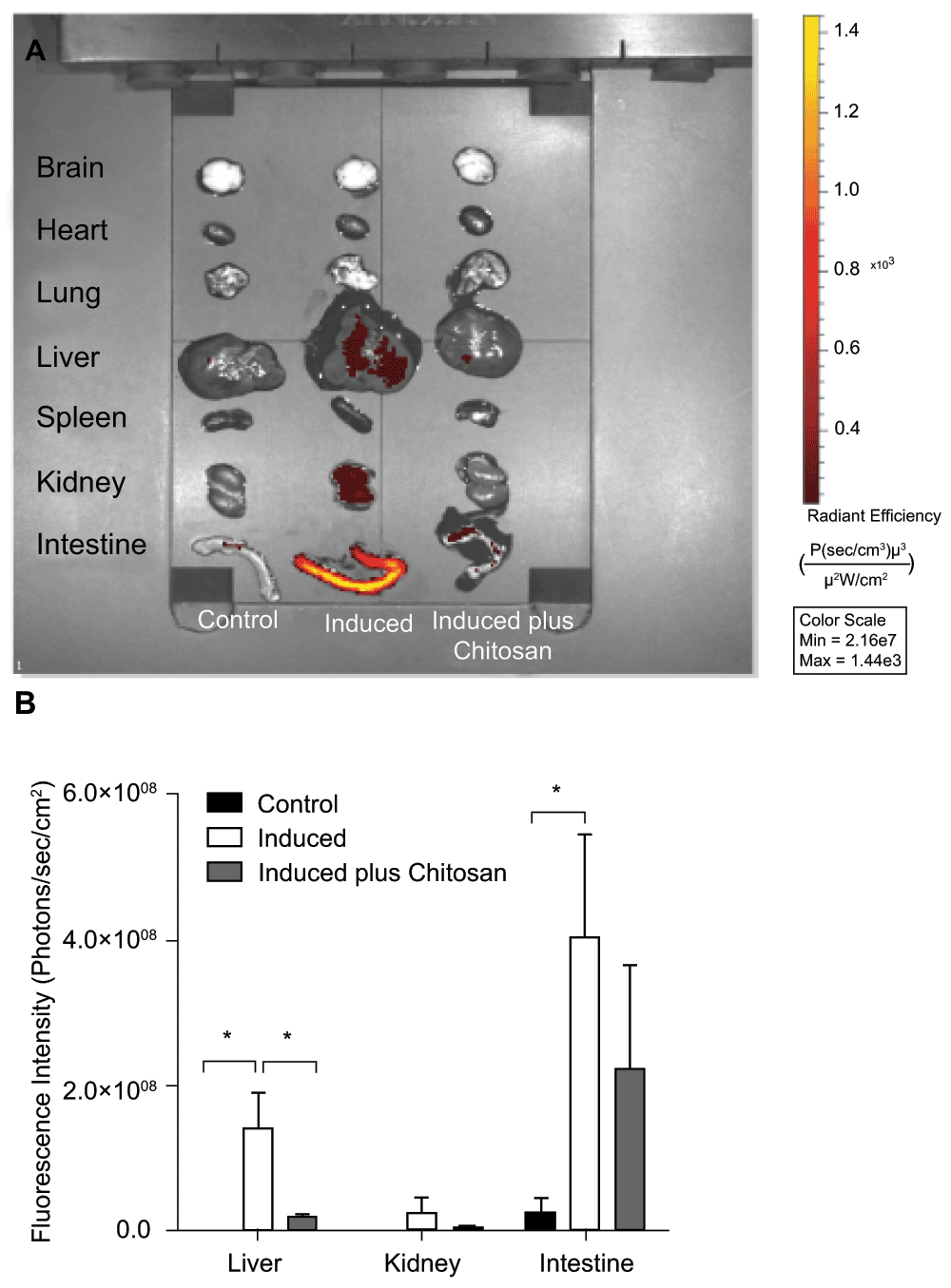
A. IVIS Representative image: ciprofloxacin was administered 2 hours post-infection to induce ϕΔTOX:GFP in vivo. A group of mice was treated with chitosan 2 hours after bacteriophage induction. All mice were sacrificed 24 hours post-infection and brains, hearts, lungs, livers, spleens, kidneys and intestines were harvested and analyzed by IVIS. Fluorescence intensity was recorded as photons/sec/cm2, and the signal intensity represents the amount of GFP present. B. Graphic of fluorescence intensity on GFP-positive organs. Four animals per group were analyzed and the fluorescence intensity was quantified using Living Imaging 4.3.1 in Calipter Life Sciences.
In order to evaluate the in vivo effect of chitosan during the infection process, mice were orally challenged with a wild-type EDL933W strain, based on the model described by Brando and collaborators9. Another mouse group was also treated with chitosan, administered orally 2 hours post-infection, and survival was followed for one week. In this preliminary study, partial protection was observed in mice treated with chitosan, resulting in a delay in the death time (Figure 4). Mice infected with EDL933W strain died at 72 hours post-infection, and mice infected followed by treatment with one dose of chitosan died at 168 hours after infection.
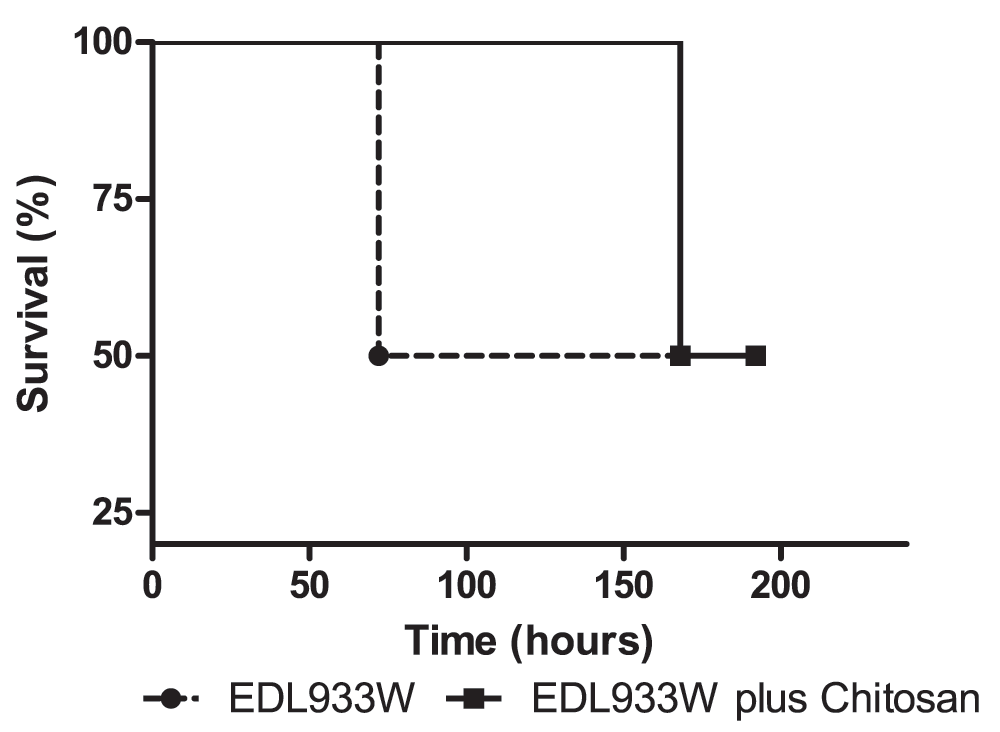
Mice were infected orally with EDL933W strain. Controls did not receive chitosan (dots and broken line) and the experimental group received chitosan 2 hours post-infection (square and fill line). Survival rates were observed for one week: two mice infected with EDL933W died 72 hours post-infection, while two mice infected with EDL933W plus chitosan died 168 hours post-infection. The remaining two mice of each group survived 192 hours post-infection.
Lambda bacteriophages are used in gene transfer and vaccine delivery because of their capacity to transduce mammalian cells in vivo10. Tyler and collaborators recently showed that prophage induction is required for renal disease and lethality in the EHEC mouse model, suggesting that free bacteriophages encoding Stx may play a direct role in the disease3.
In previous reports, we have showed that the native phage promoter controlling Stx expression is active in eukaryotic cells as demonstrated both in vitro1 and in vivo2. Based on these results and the reports previously described, we sought to evaluate whether bacteriophages could be considered a target for treating STEC infections. To this aim, we measured GFP by the strain C600ΔTOX:GFP and the mortality of infected mice following bacteriophage induction, and in vivo inactivation upon chitosan treatment positive expression was analyzed. GFP was observed in liver, intestine and kidney by IVIS on mice in which the bacteriophage lytic phase was induced by ciprofloxacin following infection. Of particular relevance was the observation that chitosan exerted a direct inactivation effect on ϕΔTOX:GFP in vitro and drastically reduced the detection of fluorescence in mice orally infected with the C600ΔTOX:GFP strain. Bacteriophage transduction of mammalian cells was also inhibited after incubation with chitosan.
Our findings indicate that chitosan possesses strong anti-bacteriophage properties in vitro and in vivo. This positively charged polymeric polysaccharide has been reported to inhibit other bacteriophages and probably acts through electrostatic interactions with negatively charged capsid proteins5. Based on these effects we propose that chitosan may be a viable alternative for the treatment of STEC infections. Chitosan is already used in food and medicine, and it is harmless to humans, making it a cheap and safe option for this application.
The fact that only partial protection was observed in vivo using chitosan may be due to its short half-life11. Our results may contribute to understand why only small numbers of bacteria are sufficient to induce HUS in humans. If bacteriophages are induced in the gastrointestinal tract, then replicate, infect bacteria in the intestine and transduce host cells, small numbers of bacteria should be enough to produce a Stx concentration sufficient to cause significant damage.
Altogether, these findings suggest a paradigm change on the role of bacteriophages in STEC infections, indicating they may be responsible for the development of disease rather than their bacterial host. Thus, prophylaxis and treatment of human bacterial infections carrying virulence factors on lysogenic bacteriophages may require targeting of the bacteriophages instead of, or as well as, the bacteria and toxins involved.
Figshare: Data sets for bacteriophage induction and effect of chitosan. http://dx.doi. 10.6084/m9.figshare.96079412
LVB designed and performed experiments, analyzed the data and wrote the manuscript; LCSF provided advice on experimental design, data interpretation, obtained funding and critical reading of the manuscript; JHA performed experiments and provided advice on experimental design, MJRR-WBL-BFMMP, PDG and RCCF provided advice on experimental design. EGS provided critical feed-back and editing on the manuscript.
This work was supported by PICT 2411 from the Agencia Nacional de Promoción Científica y Tecnológica, Argentina (to L.V.B) and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP), Brazil (to L.C.S.F). LVB and PDG are members of the Research Career of CONICET (Consejo Nacional de Ciencia y Tecnología).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We would like to acknowledge Dr. Alison A. Weiss for providing strain E. coli C600: ΔTOX-GFP.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 26 Aug 14 |
read | read | read |
|
Version 1 18 Mar 14 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)