Keywords
Function, heterologous expression, in vivo, Oryza, oxygen
This article is included in the Oxygen-binding and sensing proteins collection.
Function, heterologous expression, in vivo, Oryza, oxygen
We incorporated some of the references suggested by Dr. Matilla into the revised version of the article and indicated in the legend to Figure 1 that molecular sizes and molecular masses correspond to the Hb cDNAs and proteins analyzed in this work.
See the authors' detailed response to the review by Robert Hill
Non-symbiotic hemoglobins (nsHbs) are O2-binding proteins widely distributed in land plants, including rice1. The nsHbs are classified into type 1 and type 2 (nsHbs-1 and nsHbs-2, respectively) based on sequence similarity and O2-affinity2,3. The O2-affinity of nsHbs-1 is very high mostly because of an extremely low O2-dissociation (koff) rate constant3–5 resulting in that nsHbs-1 apparently do not release O2 after oxygenation6,7. In contrast, the O2-affinity of nsHbs-2 is moderate mostly because of a moderate to high koff rate constant for O2, thus apparently nsHbs-2 easily release O2 after oxygenation2,3,6,7. Hence, it is possible that the in vivo function of nsHbs-1 is other than O2-transport and that nsHbs-2 function in vivo as O2-carriers.
Five copies (hb1 to 5) of the nshb gene have been detected in the rice genome, which are differentially expressed in embryonic and vegetative organs from plants growing under normal and stress conditions8–11. Based on the available information on the properties of rice nsHbs and data from the analysis of other plant and non-plant Hbs, it was proposed that rice nsHbs could exhibit a variety of functions in vivo, including O2-transport, O2-sensing, NO-scavenging and redox-signaling6,12,13. However, the in vivo activity of rice nsHbs has been poorly analyzed12. An in vivo analysis for rice nsHbs is essential to elucidate the biological function(s) of these proteins. An approach to analyze the in vivo activity of nsHbs is generating knock out rice for individual nshb genes, however this is complicated because of the existence of five copies of nshb in the rice genome. An alternative approach to analyze the in vivo activity of rice nsHbs is examining individual rice nsHbs in a heterologous system, such as recombinant Escherichia coli. Rice Hb14 and Hb214 are nsHbs-1 that have been generated in recombinant E. coli TB1. The rice Hb1 and Hb2 amino acid sequence4, tertiary structure15 and rate and equilibrium constants for the reaction of O24,14 are highly similar. Thus, it is possible that rice Hb1 and Hb2 function similarly in vivo. As an initial approach to test this hypothesis we analyzed the effect of the synthesis of rice Hb1 and Hb2 in the recombinant E. coli TB1 growth. Our results showed that synthesis of rice Hb1 and Hb2 inhibited the recombinant E. coli TB1 growth and that growth inhibition was stronger when recombinant E. coli TB1 synthesized rice Hb2 than when synthesized rice Hb1.
Untransformed (wild-type) and transformed (recombinant) E. coli TB1 (Invitrogen, CA, USA) containing the constitutive pEMBL18+::Hb14, pEMBL18+::Hb214, pEMBL18+::Lba16, pEMBL18+::LbII17 and pUC18::VHb18 plasmids were grown in LB broth (Sigma-Aldrich, MO, USA) at 37°C with shaking at 200 rpm. Plasmids pEMBL18+::Lba, pEMBL18+::LbII and pUC18::VHb were included as an O2-carrier control since they code for the synthesis of the O2-carrying soybean leghemoglobin a (Lba), cowpea leghemoglobin II (LbII)17,19,20 and Vitreoscilla Hb (VHb)21,22, respectively. The existence of the VHb insert into the pUC18::VHb plasmid was verified by PCR (30 cycles at 55°C/30s for annealing, 72°C/30s for extension and 95°C/30s for denaturation) using specific oligonucleotides (VitHb/ATG: 5´-ATG TTA GAC CAG CAA ACC ATT-3´ and VitHb/TAA: 5´-TTA TTC AAC CGC TTG AGC GTA-3´) designed from the vhb sequence deposited in the Genbank database under the accession number X13516. The existence of the Hb1, Hb2, Lba and LbII inserts into the pEMBL18+::Hb1, pEMBL18+::Hb2, pEMBL18+::Lba and pEMBL18+::LbII plasmids, respectively, was verified by EcoRI- and NcoI (Invitrogen, CA, USA) -double digestion. Inserts were detected by electrophoresis in a 1.4% agarose gel. The existence of recombinant Hbs in cell soluble extracts was verified by SDS-PAGE in a 12.5% polyacrylamide gel. Evaluation of the effect of the Hb synthesis in the recombinant E. coli TB1 growth was performed in 50 ml cultures inoculated with ≈5 × 108 colony forming units from a 20 ml overnight culture. Wild-type E. coli TB1 was included as control. All assays were performed in triplicate. Cell growth was quantitated by spectrophotometry using λ = 650 nm for an 8.5 h period.
Electrophoretic analysis of the PCR reaction and EcoRI- and NcoI-double digestions showed that plasmids isolated from recombinant E. coli TB1 contained inserts corresponding to the rice Hb14, rice Hb24, soybean Lba16, cowpea LbII17 and Vitreoscilla Hb18 cDNAs (Figure 1A). Likewise, analysis by SDS-PAGE showed that rice Hb1, rice Hb2, soybean Lba, cowpea LbII and Vitreoscilla Hb existed in the soluble extracts of recombinant E. coli TB1 (Figure 1B). This evidence indicated that rice Hb1, rice Hb2, soybean Lba, cowpea LbII and Vitreoscilla Hb were synthesized by recombinant E. coli TB1.
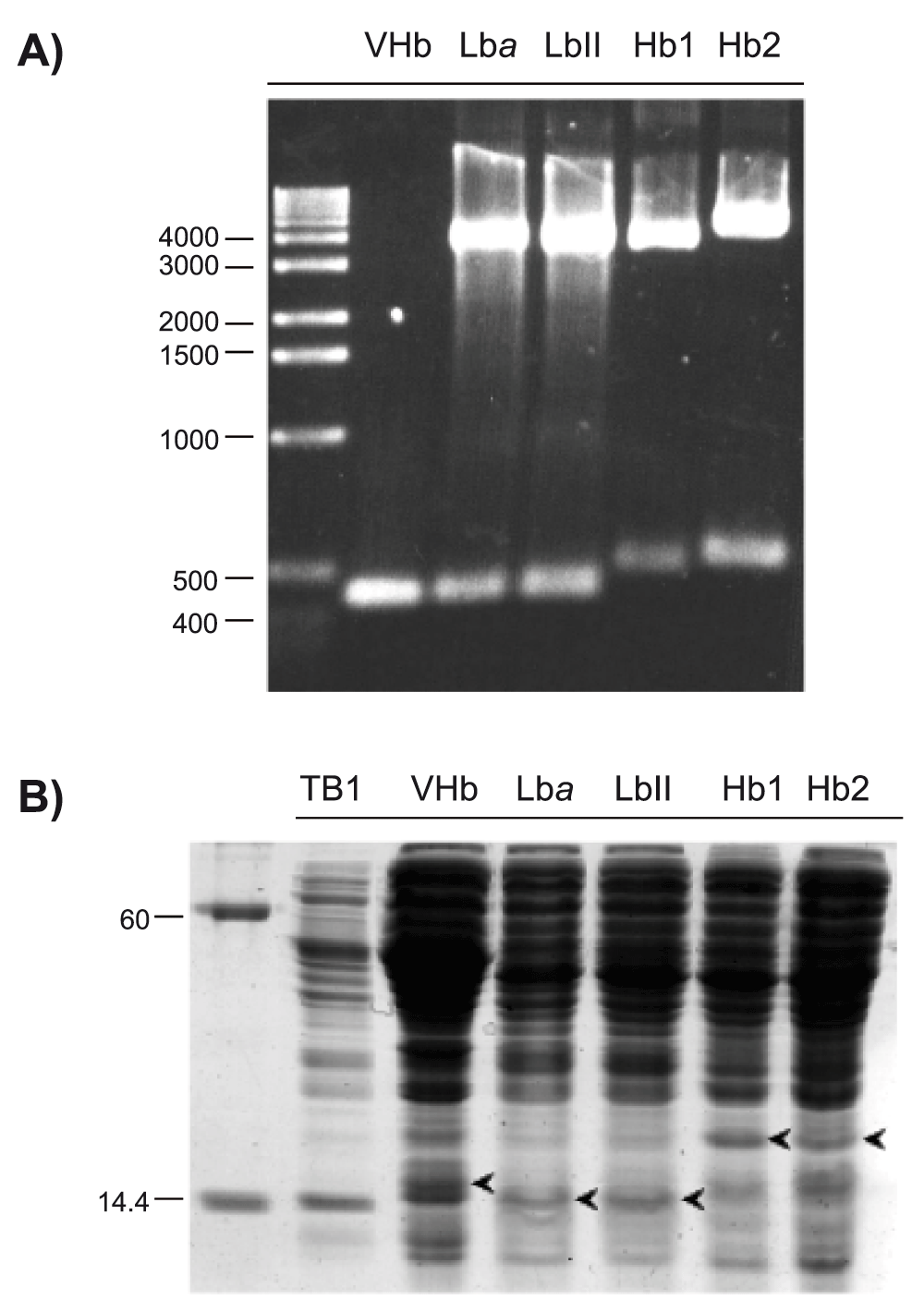
(A) Detection of Vitreoscilla Hb PCR fragment and soybean Lba, cowpea LbII, rice Hb1 and rice Hb2 cDNAs from recombinant E. coli TB1 by agarose gel electrophoresis. PCR fragment and cDNA sizes are within the 435 to 507 base pairs range, which corresponds to the molecular sizes of the Hb cDNAs analyzed here. Molecular size markers are indicated in base pairs. (B) Detection of Vitreoscilla Hb, soybean Lba, cowpea LbII, rice Hb1 and rice Hb2 proteins (arrow heads) from recombinant E. coli TB1 soluble extracts by SDS-PAGE. A 20 to 50 μg aliquot of total soluble proteins was loaded onto each lane. Recombinant Hb masses are within the 14 to 18.4 KD range, which corresponds to the molecular masses of the Hbs analyzed here. Mass markers are indicated in kD.
Figure 2 shows that synthesis of rice Hb1, rice Hb2, soybean Lba, cowpea LbII and Vitreoscilla Hb inhibited the recombinant E. coli TB1 growth. This was unexpected for soybean Lba, cowpea LbII and Vitreoscilla Hb because these proteins would promote cell growth due to their O2-transport activity17,19–22. However, under the conditions tested in this work apparently soybean Lba, cowpea LbII and Vitreoscilla Hb affected some aspects of the recombinant E. coli TB1 metabolism, possibly owed to the constitutive expression of these proteins into the host cells. Synthesis of rice Hb1 inhibited the recombinant E. coli TB1 growth similarly (∼37%) to the synthesis of soybean Lba, cowpea LbII and Vitreoscilla Hb. This observation suggests that rice Hb1 could function in vivo similarly to O2-carrying Hbs. Likewise, synthesis of rice Hb2 also inhibited the recombinant E. coli TB1 growth. However, growth inhibition was stronger (∼61%) when recombinant E. coli TB1 synthesized rice Hb2 than when synthesized rice Hb1. This observation suggests that rice Hb2 could function in vivo by scavenging O2, possibly owing to its extremely low koff rate constant for O214.
Results presented in this work suggest that in spite of the high similarity between rice Hb1 and Hb2 these proteins could function differently in vivo. In order to elucidate the apparent metabolic effects generated by the synthesis of rice Hb1 and Hb2, future work might focus on the physiological and biochemical characterization of recombinant E. coli TB1. This may include measuring cell respiratory rates and identifying cell proteins and metabolites using oximetry and proteomic and metabolomic approaches, respectively. Results from these analyses could provide valuable information to understand the in vivo function of rice nsHbs.
EAS and RAP conceived the study. EAS executed the experiments. RAP prepared the first draft of the manuscript. EAS and RAP revised the draft manuscript and have agreed to the final content.
This work was partially financed by SEP-PROMEP (grant number UAEMor-PTC-01-01/PTC23) and Consejo Nacional de Ciencia y Tecnología (CoNaCyT grant numbers 25229N and 42873Q), México, to RA-P.
I confirm that the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authors wish to express their gratitude to Dr. Dale A. Webster (Illinois Institute of Technology, USA) for kindly providing the pUC18::VHb plasmid.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 01 Mar 16 |
read | |
|
Version 1 12 Oct 15 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)