Keywords
Drosophila development, gene regulatory network, protein interaction network, visualization, web-tool, database, community annotation, ontology
This article is included in the Eye Health gateway.
Drosophila development, gene regulatory network, protein interaction network, visualization, web-tool, database, community annotation, ontology
Developmental biology is the study of processes that generate an entire organism from a single cell. A central question in this field is how differentiation produces specific cell types from pluripotent precursors. Drosophila melanogaster (D. m.) serves as a suitable and well-established model organism to address this question for numerous reasons including a short generation time, the multitude of available genetic methods, and its orthology shared with vertebrates (Reiter et al., 2001). The D. m. eye allows the study of morphological rearrangements as well as differentiation of non-neuronal and neuronal cell types like photoreceptors (PRs) on the single cell level (Thomas & Wassarman, 1999).
Understanding cell differentiation requires knowledge of the involved genes, their temporally varying (dynamic) expression patterns, and interactions. Interaction data from different sources are accessible through e.g. Biogrid, Intact, String, and REDfly (Chatr-Aryamontri et al., 2015; Gallo et al., 2011; Orchard et al., 2014; Szklarczyk et al., 2015) and database collections, e.g. iRefindex or mentha (Calderone et al., 2013) (Razick et al., 2008). Expression data is mostly provided on embryonic development or organ systems, e.g. by FlyBase (dos Santos et al., 2015) and the Berkeley Drosophila Genome Project (BDGP) (Hammonds et al., 2013), but the coverage and precision of expression pattern annotation on the cellular level and the temporal resolution at later stages, e.g. larva and pupa, are limited (see Supplementary Material). On the other hand, a wealth of expression pattern data on these levels is contained in publications (e.g. (Potier et al., 2014)). Systematic use of these data requires their structured assembly through an extensive curation effort. Since automated curation is prone to errors (Mao et al., 2014), information must be extracted manually from the literature by experts, who can critically interpret the respective types of data like micrographs or expression profiles (Tomancak et al., 2007) and convert these to machine-readable data for further computer-based analyses.
We have thus developed FlyOde, an online hub for the community-driven systematic assembly of data on D. m. eye development. FlyOde is a web interface for interactive exploration of gene and protein relationships by combining visualization of the curated gene regulatory network with filters specific to fly development. FlyOde is built on an ontology-driven curation process that stores data in a specifically formatted text file, which can easily be enriched and extended upon arrival of new data.
A directed gene interaction network representing eye development of D. m. from the third instar larva to the adult with focus on PR differentiation was constructed using Cytoscape (Shannon et al., 2003), based on data extracted manually from 77 publications (Figure 1). Currently, the network contains 146 nodes representing genes/proteins, and 284 edges representing activating or inhibiting genetic, protein-protein, or protein-DNA interactions. The layout is generated manually and organized to approximately represent developmental time along the horizontal axis, beginning with early third instar larval stage on the left, and network hierarchy along the vertical axis with master regulators (as defined by (Chan & Kyba, 2013)) placed towards the top. Nodes are associated with their FlyBase symbol, name, alternative names, FlyBase link, dynamic expression pattern, phenotypes, the terms for each of the three Gene Ontologies (GO) (Ashburner et al., 2000), and the literature references. GO annotations were added using Cytoscape. Expression pattern annotation follows a specifically developed ontology which links developmental stage and cell type (Figure 2). We support and encourage annotation from the community to continually extend the dataset.
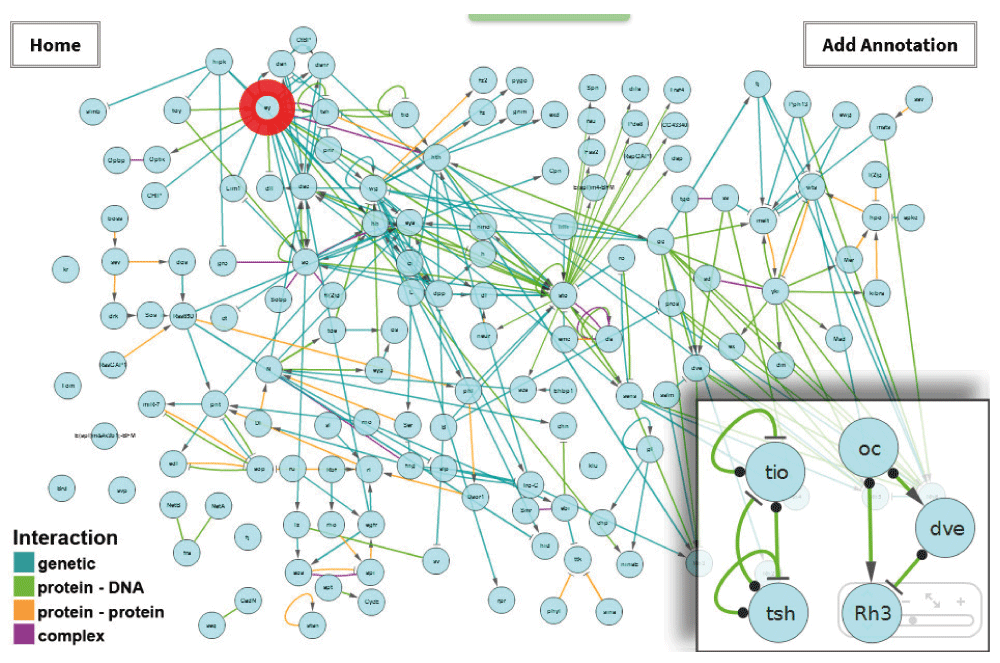
The red circle indicates the eyeless gene (see Example 1). The inset shows selected interactions containing network motifs: negative autoregulation and feedback loop (left) and an incoherent feed forward loop (right).
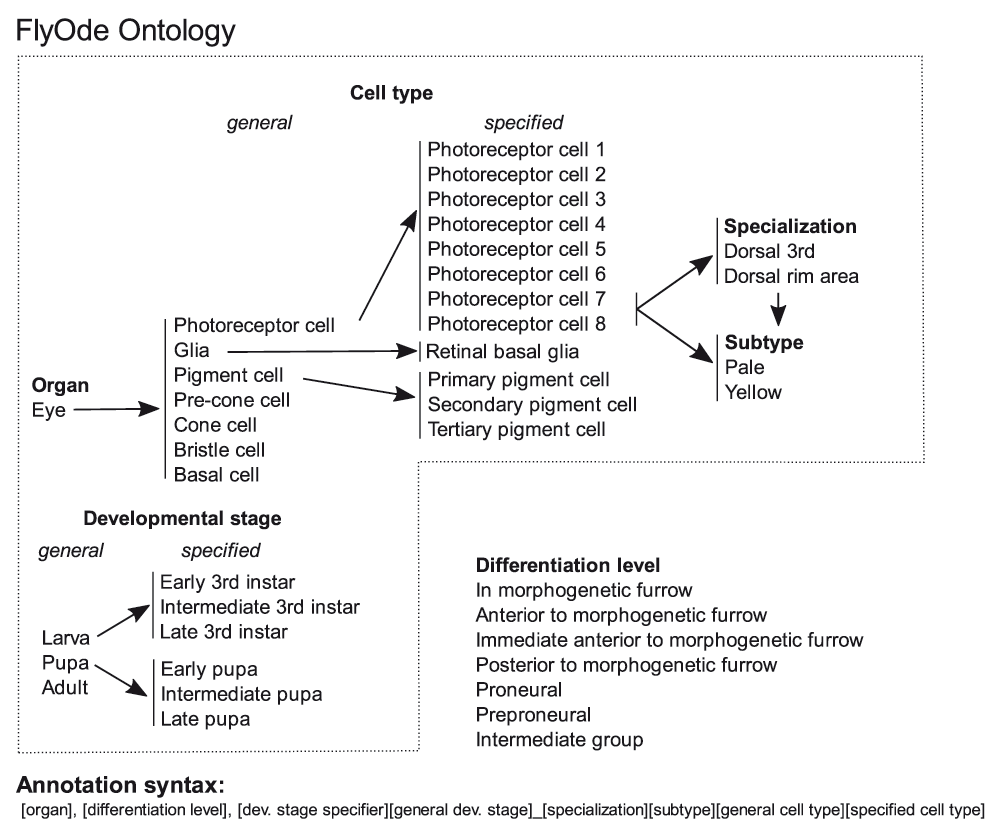
Hierarchical representation of controlled vocabulary for the gene annotation with dynamic expression patterns is shown. In the annotation syntax terms for cell type and developmental stage are directly linked. Each term-combination represents expression in a specific cell type and developmental stage. As many comma-separated term combinations as needed to describe the expression pattern are given. During retinal development of Drosophila, cells at consecutive stages of differentiation coexist at the same developmental stage of the organism. The morphogenetic furrow marks the initiation of differentiation and moves across the eye precursor organ, the eye imaginal disc. Thus, the expression pattern is also defined by the localization relative to the morphogenetic furrow. This and additional terms that are common in the field are assigned to what we call “differentiation level”.
The interactive JavaScript based web application renders the manually curated network, which is embedded using Cytoscape Web (Lopes et al., 2010). Genes are represented as nodes and gene (or protein) interactions are visualized as directed edges. Nodes and edges have been annotated during the curation process (see previous section), and this information is displayed when an item is selected. For the selection of nodes a module for searching gene symbols, names, alternative names, and GO-terms is provided. Additionally, the network can be filtered according to gene expression patterns using dropdown menus whose structure follows the FlyOde ontology (Figure 2). Filters can be combined with the Boolean operators AND, OR, and NOT. Extensions to the network can be submitted to us via a community curation form.
To characterize the content of the current FlyOde network GO enrichment analysis was performed with the ClueGO app in Cytoscape (Bindea et al., 2009). It shows that terms correlated with development and morphogenesis, pattern formation and polarity, apoptosis, mitosis, and regulation of transcription are highly over-represented, as expected (see Supplementary Materials and Supplementary Figure 1 and Supplementary Figure 2).
To evaluate the information that can be obtained from FlyOde the FlyOde filter function was compared with the FlyBase QueryBuilder. When queried for general terms like “larva and PR”, FlyBase gave more gene hits than FlyOde, but when increasingly specifying the developmental stage and cell type, like “pupa and dorsal rim area R8 (photoreceptor cell 8)”, more genes were found with FlyOde as compared to FlyBase. This shows that FlyOde already stands out in defining gene expression in a specific cell type and during a specific developmental stage of PR differentiation (see Supplementary Materials and Supplementary Figure 3).
To get an idea of the basic mechanisms represented in FlyOde, the occurrence of network motifs was analysed manually in Cytoscape. Motifs commonly found during development of organisms, like autoregulation, feedback and feed forward loops were observed. This indicates that FlyOde at least partially displays the connectivity and level of detail to qualify for representation and detailed analysis of developmental processes (see Figure 1 and Supplementary Materials, Supplementary Figure 4 and Supplementary Table) (Alon, 2007; Davidson, 2010).
In this example we use the search functionality to explore a specific gene in the dataset. In order to do so, Pax6 is entered into the search box. This gene is known under that name as an important regulator of development in many organisms (van Heyningen, 2002). However, the official name in D. m. is eyeless. Due to its annotation with alternative names it is found despite the search for the unofficial name Pax6 and highlighted in the network (Figure 1). Its position on the left side of the graph indicates that it is mainly expressed early during eye development. In the web interface additional information is displayed in a content related text field below the graph, which we designate “report panel”. A closer look at the expression pattern displayed there shows that it is expressed anterior to the morphogenetic furrow, in all photoreceptors in the early third instar larva, and in outer photoreceptors in the late pupa and adult. The top position in the hierarchy indicates that it is a master regulator. This is supported by its high number of interactions, and by the phenotypes, which are given in the report panel (Chan & Kyba, 2013). We also find that GO annotates eyeless as a transcription factor, and that it is involved in developmental processes of other organs. Finally, the literature references and the FlyBase link can be followed for further information.
Here we apply filter combinations to display genes that are expressed in R8, but not in any other PR in the intermediate pupa to obtain candidate markers for R8 in immunostaining and cell sorting experiments. In the dropdown menus we choose “pupa”, “intermediate pupa”, “photoreceptor cell”, and “R8”, respectively. We add another filter line with the Boolean “NOT”, and in the dropdown menus select “intermediate pupa”, “photoreceptor cell”, “R7”, and another filter line with the Boolean “NOT”, “intermediate pupa”, and “at least one outer PR”, which in combination with “NOT” means “no outer PR”. All nodes are removed except for scabrous and senseless, which are therefore candidates for being R8 markers. The literature references for these two markers provide a starting point for future experimental studies.
More examples are provided in the tutorial at http://flyode.boun.edu.tr/quickguide.html.
Here we have presented FlyOde, which provides a platform for combining published data on gene regulatory networks (GRNs) of Drosophila organ development. As a starting point, we have equipped FlyOde with extensive GRN data for D. m. eye development, such that the web interface serves as a versatile tool for everyday tasks a fly researcher encounters.
FlyOde delivers high quality data standards by manual curation. We expect to achieve efficient data collection by distributing the annotation workload among community members with minimal effort for the individual contributors, who only need to submit a simple form to add new nodes, interactions, or annotation. In return, they directly profit from the improved tool by linking their data of interest with the shared knowledge.
We are constantly improving the ontology and web application, in parallel to ongoing data curation and dataset extension. FlyOde will be expanded to include other organs with the ultimate goal to compare their GRNs.
Future work will profit from the obtained data by constraining network inference from gene expression data (Hecker et al., 2009). Another anticipated approach is to assign quantitative expression data to the established network to facilitate mathematical modelling (Graham et al., 2010).
We envision FlyOde as a companion that guides researchers through developmental processes, for example while studying a paper, and believe that the community-curated dataset and its analysis will add significant knowledge to developmental biology.
FlyOde, including all network data, annotations and their corresponding references can be freely accessed via the web-application at http://flyode.boun.edu.tr/.
The source code for the web application can be downloaded from Github (https://github.com/begum-alaybeyoglu/FlyOde).
S.A.K. established the concept, network, annotation and ontology, performed analysis and wrote the manuscript. B.A. generated the web application. C.X.W. performed analysis and provided input during manuscript preparation. A.C. coordinated the project. All authors contributed to proofreading the manuscript and have agreed to the final content of the manuscript.
S.A.K. was supported by TÜBITAK 2236 Co-Funded Brain Circulation Scheme.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Stefan Fuss and the Bogazici University Computer Centre for help with the server and Elif Özkirimli (all Bogazici University, Istanbul) for proofreading the paper.
Supplementary Figure 1.
GO-term enrichment analysis with ClueGO.
Click here to access the data.
Analysis and characterization of the FlyOde network.
Includes Supplementary Figures 2, 3, 4.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 21 Dec 15 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (1)