Keywords
Zika, virus infection, protein interaction, interferon-inducible transmembrane proteins
This article is included in the Emerging Diseases and Outbreaks gateway.
This article is included in the Zika & Arbovirus Outbreaks collection.
Zika, virus infection, protein interaction, interferon-inducible transmembrane proteins
The Zika virus (ZIKV) is a flavivirus that was initially isolated from rhesus monkeys in 1947 and was first reported in humans in 19521. Until recently, reports of this virus had been limited to Africa and Asia2 but currently there is an ongoing, wide-spread Zika epidemic3. The virus has rapidly spread across the Americas and has been declared a ‘global emergency’ by the World Health Organization4. It is mostly transmitted by mosquitoes and clinical manifestations include rash, mild fever, arthralgia, conjunctivitis, myalgia, and headaches. In addition, it has been reported recently that the virus can be transmitted sexually, with the risk of infection persisting for several months after initial contact5. Earlier, the symptoms of ZIKV had been reported to be mild1, but, the virus has been recently linked to two more serious afflictions: Guillen-Barré syndrome6,7 and microcephaly8–12, both of which are serious neurological conditions. Microcephaly results in reduced head circumference measurement in infants, exhibiting complications in brain development. Of particular concern is the attribution of microcephaly to infection with ZIKV occurring between the first two trimesters of pregnancy11,12. Evidence linking ZIKV to microcephaly includes detection of ZIKV RNA in tissue such as the placenta and amniotic fluid of pregnant women with ZIKV, as well as in the brains of stillborn infants with microcephaly13. In a study with human induced pluripotency stem cells, the mechanism of ZIKV related cell death has been elucidated. This study demonstrated that ZIKV infects human embryonic cortical neural progenitor cells (hNPCs), ultimately leading to attenuated population growth mediated by virally induced caspase-3-mediated apoptosis and cell-cycle dysregulation14. Mice studies showed that ZIKV infection can lead to nerve degeneration, softening of the brain and porencephaly15.
Very recently, two small membrane-associated interferon-inducible transmembrane proteins (IFITMs) IFITM1 and IFITM3 were discovered to have a protective role against the Zika virus infection by inhibiting replication of the virus and preventing cell death induced by Zika virus5. IFITMs were shown to have an inhibitory role against other flaviviruses also, such as West Nile and dengue virus. Type 1 interferon (IFN) signaling inhibits Zika virus pathogenesis. Prior to induction of IFN-stimulated genes, IFITMs may provide initial defense against the infection5. However, since the exact mechanism of IFITM1 and IFITM3 mediated restriction are yet unknown, computational methods could accelerate research by presenting testable hypotheses.
In our earlier work, we developed a computational model called ‘High-confidence Protein-Protein Interaction Prediction’ (HiPPIP) model that identifies novel protein-protein interactions (PPIs) in the human interactome16, motivated by the fact that PPIs prove to be valuable in understanding the function of a gene, and specifically in how it plays a role in causing or preventing disease. One example of the impact of these computational predictions is the PPI that we predicted between OASL and RIG-I16, which was validated to be a true PPI through co-immunoprecipitation16,17. This led to the formulation of a hypothesis about its significance and led to the discovery of its functional relevance, namely that upon viral infection, OASL triggers the immune system by activating the RIG-I pathway, thus inhibiting virus replication18. functional studies initiated solely by this predicted PPI showed that human OASL binds to dsRNA to enhance RIG-I signaling, and that boosting OASL can help inhibit viral infection18. In this work, we applied HiPPIP model to discover novel PPIs of IFITM1 and IFITM3, to potentially accelerate the discovery of the mechanism by which they inhibit ZIKV and other viral infections.
PPIs were assembled by collecting known PPIs from the Human Protein Reference Database (HPRD)19 and Biological General Repository for Interaction Datasets (BioGRID)20, and by computing novel PPIs using the HiPPIP model that we developed16. Computationally discovered PPIs have been shown to be highly accurate by computational evaluations and experimental validations of a few PPIs16. Interactome figures were created using Cytoscape21. Pathways associated with proteins in the interactome were collected using Ingenuity Pathway Analysis® suite (http://www.ingenuity.com). Gene Ontology terms enriched among the interacting partners (including the candidate genes IFITM1 and IFITM3) were computed using the BiNGO plugin of Cytoscape22.
We assembled the PPIs of IFITM1 and IFITM3 (Figure 1) by computing novel PPIs using HiPPIP model and collecting known PPIs from publicly available databases, Human Protein Reference Database (HPRD) and Biological General Repository for Interaction Dataset (BioGRID)23,24. We found that both proteins have known PPIs with proteins involved in immunity, and several novel (predicted) PPIs with proteins that seem to have relevant functions. DEAF1 is involved in neural tube closure, embryonic skeletal development and anatomic structure morphogenesis, and other functions. FNDC3B was found to be associated with heart rate, height and corneal structure through genome-wide association studies. It is a membrane protein, and, while its own functions are unknown, its interactors are involved in regulation of glial cell apoptotic process, regulation of ion transport (sodium, potassium, calcium) and several cardiac processes. SPTA1 is involved in neural functions of actin filament organization, neurite outgrowth and axon guidance. RASSF7 is localized to microtubule organizing center. While its function is unknown, it interacts with proteins that are involved in cell proliferation in brain, regulation of neuroblast proliferation, nervous system development, synaptic vesicle fusion to presynaptic membrane, and viral budding and assembly. TSSC4 interacts with both IFITM1 and IFITM3. TSSC4’s functions are unknown but its own interactions suggest that it may be involved in viral penetration into host nucleus, protein import into nucleus and immune response signaling, among other processes. TLR7 is involved in several functions and pathways related to innate immunity. ARPC1B is part of actin related protein 2/3 complex; its interactions suggest that it may be involved in neuronal development such as axonogensis and development, neuron differentiation, nervous system development, and immune related terms such as innate immune response, regulation of immune response, etc. These functional annotations are sourced from Schizo-Pi16,25; for example, see: http://severus.dbmi.pitt.edu/schizo-pi/index.php/gene/view/10522.
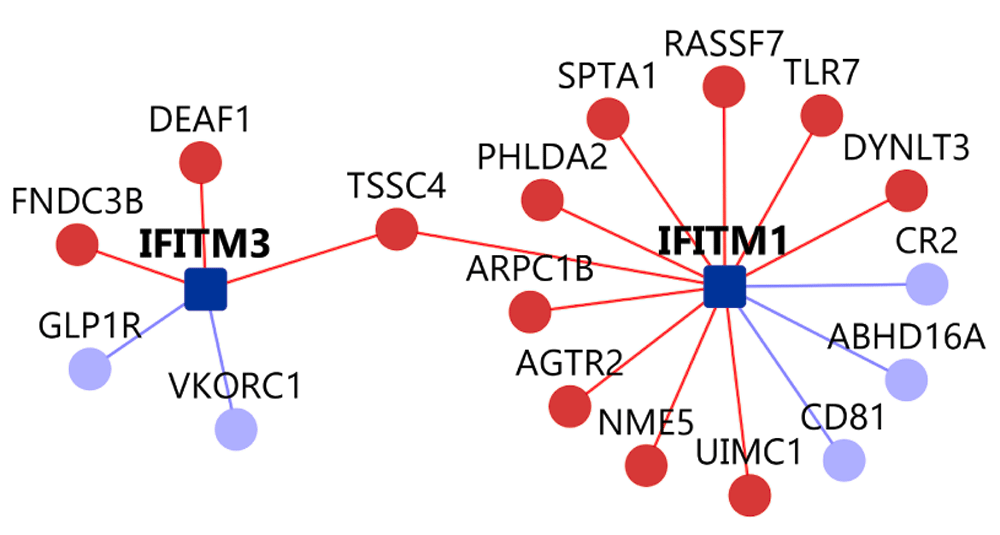
Novel interactors of IFITM1 and IFITM3 are shown as red colored nodes while previously known interactors are shown as light blue colored nodes. Novel interactors of IFITM1 and IFITM3 are shown as red colored nodes while previously known interactors are shown as light blue colored nodes.
Pathways associated with IFITM interactome computed with Ingenuity Pathway Analysis Suite® are given in Table 1. Gene Ontology biological process terms associated with the interactome, compiled with BiNGO22 are shown in Figure 2 and Table 2.
Pathway associations were computed with Ingenuity Pathway Analysis Suite ®. Novel interactors are shown in bold.
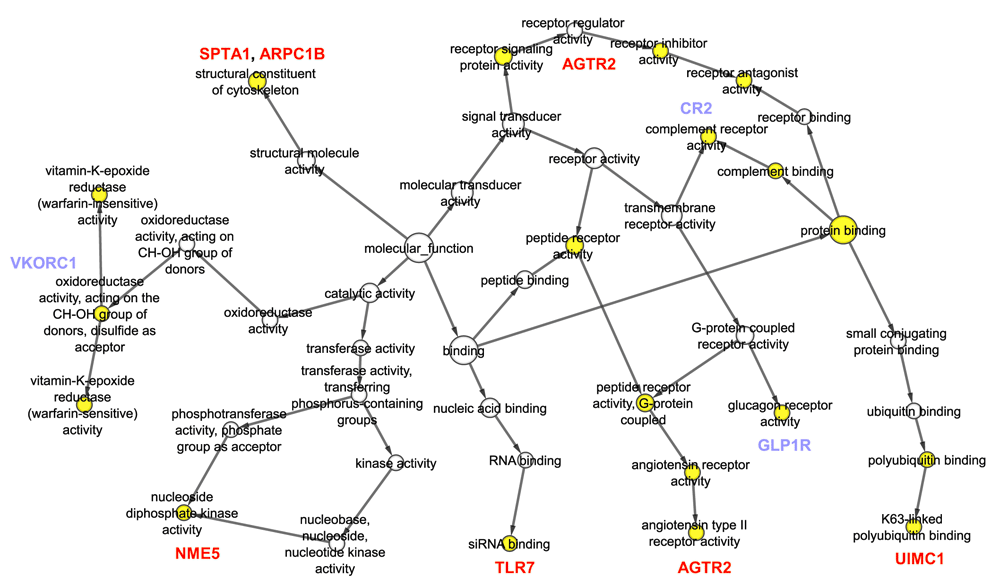
Yellow color signifies statistically significant enrichments. Novel interactors that are associated with the GO terms are shown in red and known interactors in blue. See Table 1 for a complete list of terms associated with the genes.
Novel interactors are shown in bold.
There is only one study that presents altered gene expression under ZIKV infection available in Gene Expression Omnibus14. The study with eight samples (four infected and four control samples) showed that the infection of human neural progenitor cells (hNPCs) with the virus caused increased cell death and cell-cycle dysregulation14. We examined whether any of the interacting genes were differentially expressed in that study and found five genes that were differentially expressed with a small fold-change but with significant p-value (< 0.005) (Table 2): CD81, NME5, and RASSF7 were found to be under-expressed and FNDC3B and UIMC1 were found to be over-expressed (Table 3).
See http://severus.dbmi.pitt.edu/schizo-pi for annotations of individual proteins that are compiled from various databases. Also see the following link to our LENS webserver, where we present annotations of all the genes in the IFITM1-IFITM3 interactome and also annotations of proteins that further interact with interactors (i.e. 2nd level connectors of IFITMs). Under each tab, ‘candidate genes’ refers to IFITMs and their interactors shown in Figure 1 of the paper, while entire interactome includes all of their interactors. Note that the database behind LENS does not include novel protein-protein interactions; therefore, they are not shown as edges in the network diagram. The sources of the pathways and disease associations shown on this website are given in 25,26.
http://severus.dbmi.pitt.edu/LENS/index.php/results/view/57649c2516f9a/admin_57649c251737d
All pertaining data are provided in the manuscript.
This work is funded by Biobehavioral Research Awards for Innovative New Scientists (BRAINS) grant (R01MH094564) from National Institute of Mental Health of National Institutes of Health of USA.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
MKG thanks her students Josefina Correa Menendez, Mara Staines, Varsha Embar and Srilakshmi Chaparala for assisting in this work.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
References
1. Zhu J, Zhang Y, Ghosh A, Cuevas RA, et al.: Antiviral activity of human OASL protein is mediated by enhancing signaling of the RIG-I RNA sensor.Immunity. 2014; 40 (6): 936-48 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
|
Version 2 (revision) 21 Nov 17 |
read | read | read | |
|
Version 1 05 Aug 16 |
read | read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)