Keywords
Ctenophora, Phylogenetic reconstruction, Ribosomal subunits, Non-fluorescent protein (GFP-like), Bayesian Inference, Maximum Likelihood, Isopenicillin-n-synthase (IPNS)
This article is included in the Phylogenetics collection.
Ctenophora, Phylogenetic reconstruction, Ribosomal subunits, Non-fluorescent protein (GFP-like), Bayesian Inference, Maximum Likelihood, Isopenicillin-n-synthase (IPNS)
We revised the manuscript and performed the following changes according the suggestions made by the referees for the last version of the manuscript:
See the authors' detailed response to the review by Steven H.D. Haddock
See the authors' detailed response to the review by Martin Dohrmann
Several phylogenetic hypotheses of the phylum Ctenophora based on morphological data1, ribosomal markers2,3, protein-coding markers4, have been proposed, all of them through different approaches.
Due to the poor fossil record of the group, the morphological data of fossil taxa have been not enough to help to resolve this question, because it’s impossible to determine which characteristics arose first between the ctenophores, for several reasons such as the poor conservation state of the available fossils5,6 or the lack of shared characteristics between the extant ctenophores and the extinct ctenophores7. Also some morphological characteristic have been demonstrated to be homoplastic1,3. These situations slow down the process of reconstructing the phylogeny of this phylum.
There are also difficulties establishing an appropriate outgroup, due to the unknown position of this phylum inside Metazoa, many hypotheses have been suggested8–11. This uncertainty could bias the phylogenetic analyses if a distant outgroup is chosen (eg. highly saturated sequences, not homologous sequences available, etc), affecting directly the support values and the topology of the reconstructed tree12.
In this study we reconfirm the paraphyly of order Cyddipida similarly to all previous studies, also we confirm the order Lobata is paraphyletic, the reasons are exposed in the results section. Nevertheless, our data don’t support the paraphyly of Beroidae. Due to the fairly wide taxonomic sampling of this study resulting from the fusion of the protein-coding and ribosome data, some interesting relationships are suggested, such as the placement of Cestida and Thalassocalycida orders inside Lobata.
The ribosomal sequences were obtained from public data available on GenBank and automatically downloaded, then they were classified using python scripts. The NFP sequences was also obtained from GenBank and for certain taxa supplied by Steve Haddock via e-mail, from the Supplementary data of the reporting study of the marker4, we only included sequences from Ctenophora. The Tyrosine aminotransferase sequences and the HLH domain-containing protein were also obtained from public data of GenBank. The accession numbers of the sequences used during this study are presented in Table 1.
Previous of the concatenated final analysis, we tested several markers such as:
1. Ribosomal markers: 18S, 5.8S, 28S, ITS1, ITS2
2. Non Fluorescent Protein (NFP)
3. Tyrosine aminotransferase
4. HLH domain-containing protein
We execute a single locus analysis for all these markers. The ribosomal sequences were aligned by MAFFT v.7.713 with the option –auto. The proteing coding sequences (NFP, Tyrosine aminotransferase and HLH domain-containing protein) were aligned using RevTrans2 (http://www.cbs.dtu.dk/services/RevTrans-2.0/web/)14.
Models for single locus analyses were selected with two programs: jModelTest 2.1.1015 for nucleotide datasets (ribosomal data), and ProtTest v 3.4.2 for protein markers16.
Single locus analyses were performed by partitions obtained with Gblocks 0.91b (http://molevol.cmima.csic.es/castresana/Gblocks_server.html)17,18.
For phylogenetic reconstruction using the concatenated ribosomal dataset the pipeline PhyPipe was used19 (available at: https://gitlab.com/cibiop/phypipe/).
The Bayesian inference analyses (BI) were all performed by MrBayes 3.2.620 and the Maximum Likelihood (ML) analyses were performed by GARLI 2.0121 and RAxML 8.0.022.
NFP was previously introduced as an ortholog by 4, which function is still unknown. So we took this marker along the ribosomal data as the backbone of the alignment matrix (and the study).
Concatenation of the sequences for the study (NFP,HLH,Tyr, 18S, 5.8S, ITS1, ITS2) was performed by 2matrix 1.0 (https://github.com/nrsalinas/2matrix)23. This script allows the automatic concatenation of a heterogeneous matrix, and also convert the concatenated matrix to the files input of the Maximum Likelihood and Bayesian Inference programs.
Partition and selection of the models for the concatenated matrix were performed by PartitionFinder224, separately from Ribosomal data and protein-coding markers. The best scheme files are available inside the Supplementary data.
The models used for protein-coding markers in all analyses are: For NFP, JJT+I+G25; for Tyrosine amino-transferase, LG+G26, and for HLH, VT+I27.
The set of parameters for Bayesian Inference analysis are reported in the Supplementary data inside the NEXUS file, this analysis were performed in CIPRES28, we used 8 MCMC with 10’000.000 generations by duplicate, this allows an optimal performance of the analysis. For 18S and 5.8S the analysis was performed with HKY+I+G29, for ITS1 and ITS2 with SYM+G30.
RAxML analysis was performed in CIPRES28 with 20 independent maximum likelihood analyses and 10.000 bootstrap iterations (pseudoreplicates) for nucleotide partitions, and the model used in this analysis was GTR+G+I31. The importance of invariant proportion executing an analysis with RAxML in this specific dataset is explained in Discussion section.
GARLI analysis was performed with 10 independent maximum likelihood analyses and 1004 bootstrap iterations (pseudoreplicates) for nucleotide partitions we used more specific models. For 18S we used TrN+I+G32; for 5.8S, TIMef+I+G; for ITS1, TIMef+G, and for ITS2, TVMef+G. These models were indicated by jModeltest 2.1.1015.
The models were all selected by BIC criterion. In this study we did not used an outgroup.
In order to obtain a more complete matrix, we fused sequences from few species of the same genus into a single record, for example, we fused Hormiphora plumosa and Hormiphora californiensis into a single Hormiphora sp.; Bolinopsis sp. and Bolinopsis infundibulum into a single Bolinopsis sp. and Lampea lactea and Lampea pancerina into a single Lampea sp. These fused species allow an improvement of the alignment matrix and the phylogenetic reconstruction. Also few species were duplicated, such as Beroe forskalii, Bathyctena chuni, Hormiphora sp., because the HLH marker presented variation amongst the species and was not possible to obtain a consensus. We confirm the monophyly of this variation through a single locus analysis, as mentioned before.
According to 3 partitions obtained by Gblocks did not improve the analysis for ribosomal markers, also it did not improve the analysis for Tyrosine aminotransferase. On the other hand, for NFP and HLH domain-containing protein the bootstrap values and posterior probability improved with partitioned analysis (see Supplementary data), unfortunately this marker didn’t improve the final alignment matrix results.
We found that trees reconstructed using 28S, IPNS, and the other domain-containing proteins, presented several incongruences between them and the other markers. Ribosomal markers, Tyrosine aminotransferase, Non-fluorescent protein and HLH domain-containing protein did not present any strong incongruence amongst them. For that reason those markers were chosen for the concatenated analysis (protein sequences + nucleotide sequences).
The tree reconstructed from the combined dataset (protein + ribosomal DNA) is presented on Figure 1. The results from both Maximum Likelihood analyses (RAxML and GARLI) for the combined dataset (protein + ribosomal DNA) are similar, except in the specific relationships between Eurhamphaeidae + Cestida + Leucotheidae + Bolinopsidae. RAxML results matches with Mr-Bayes results, but three nodes of the analysis have low posterior probability (BI) or low bootstrap values (ML). RAxML analysis shows a clade composed by Eurhamphaeidae and Leucotheidae, and other clade composed by Cestida and Bolinopsidae. Whereas GARLI analysis shows Eurhamphaeidae as sister taxa of Leucotheidae, Cestida and Bolinopsidae. RAxML results are similar to 3. All of our analyses show Cestida within Lobata with high bootstrap values and posterior probability, defining Lobata as a clade composed by Leucotheidae, Eurhampaheidae, Bolinopsidae and Ocyropsidae. Bathocyroidae and Lampoctenidae families have an uncertain position between Lobata and the clade composed by Beroe sp. and Haeckelidae.
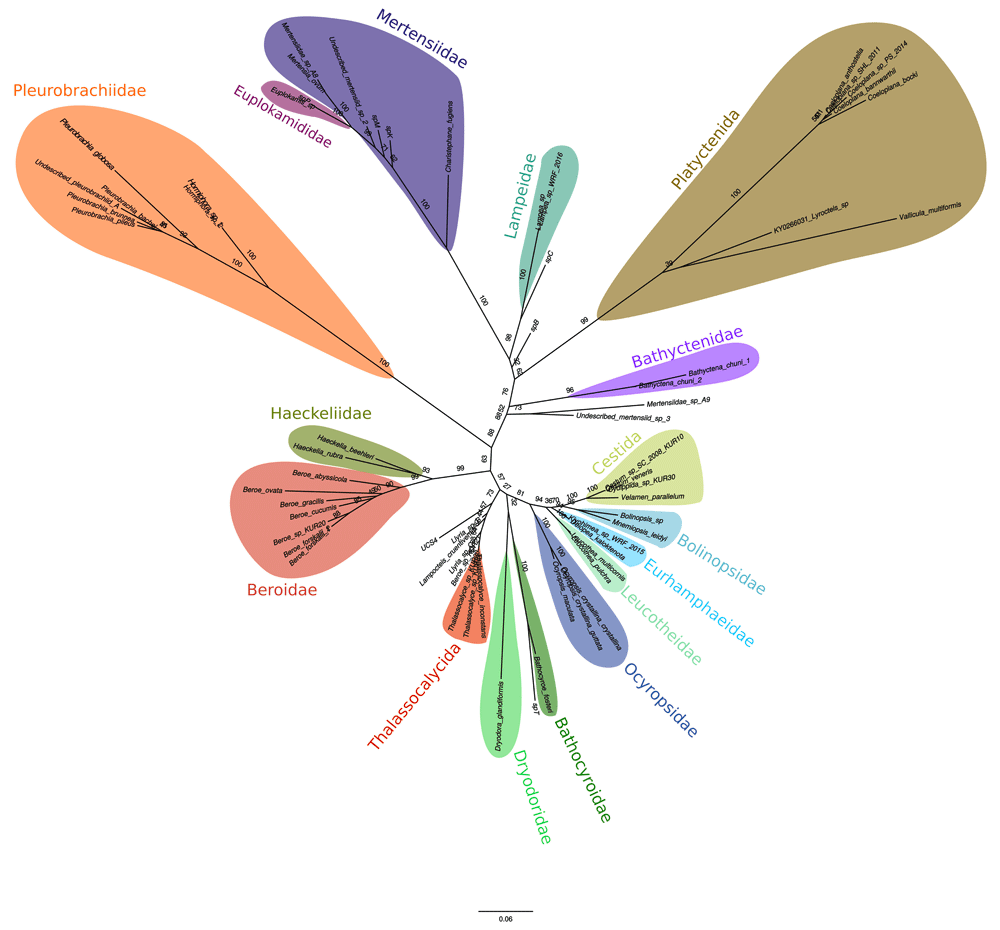
Support values of nodes (bootstrap values) are shown on tree branches.
Our analysis support a clade including Thalassocalycida and Lampoctenidae but the position of this clade remains controversial due to the lack of high bootstrap values. The family Bathocyroidae forms a clade with Dryodoridae family, but this clade has a low posterior probability and low bootstrap values, so for now it is not accurate to set hypothesis around this result, this results is similar to obtained by 4. Also the position of Dryodora glandiformis is still undetermined, in ML analysis this family could group with even Pleurobrachidae, due to the low bootstrap values of nodes between Lobata and Pleurobrachidae and with BI analysis with all lobates. Undescribed species T forms a good supported clade with Bathocyroidae. Further studies may focus in describing this taxon for morphological purposes. We executed a rogue taxa analysis through RogueNaRok33, we found that Beroe ovata, Beroe cucumis, Beroe gracilis, Lampocteis cruentiventer, Dryodora glandiformis, UCS4, Llyria B, and Lyrocteis sp. were rogue taxa during this analysis, the low bootstrap values could be related to this.
The relationship of Bathyctenidae family (Represented by Bathyctena chuni) with the Mertensiidae family and the Platyctenida order remains unclear, this family shows affinity to this clade, also in several times forms a clade with two undescribed Mertensiids (A9 and undescribed sp3), this two taxa are excluded of the family Mertensiidae (Represented by Mertensia ovum, Charistephane fugiens).
The identity of spB remains unclear. species spC forms a good supported clade with Lampeidae by both methods.
In this study, we do not include an outgroup, as consequence of it, we used mid-root point method for rooting topologies using Figtree v.1.434. Rooted trees for each analysis (RAxML, GARLI and MrBayes) are in the Supplementary material section.
By Mid-root point method, the topologies were splited in two major clades, one composed of Lyroctenidae+Coeloplanidae+Mertensiidae+Lampeidae+ Bathyctenidae and one composed of Pleurobrachiidae+Haeckelidae+Beroe sp.+Cestida+Lobata+ Dryodoriade+Thalassocalycida. These results are similar to 3. Both major clades with high bootstrap values(RAxML, 90 for both clades and GARLI, 84 for both clades) and for Mrbayes 0.96 as posterior probability for both clades.
This study also present interesting similarity to morphological study presented by Ou in 7, were the extant ctenophores (Excluding Beroida) are splited into two major clades, in one of the clades, Cyddipida+Platyctenida and other clade presenting Lobata+Cestida+Thalassocalycida+Ganeshida. Setting the paraphyly of Cyddipida would be interesting improve this morphological study, due to the similarities (until certain point) that the study present with the current. Also7 study present Beroe as the most basal inside the extant Ctenophora, this study, as previous3, denies the basal position of Beroida, also denies the Beroida as a paraphyletic group3.
Next steps for the resolving of the phylogeny of this group is to determine who is the most basal branch inside the Ctenophora, making possible and reconstruction of ancestral characters. Inside the upper clade formed by Pleurobrachiidae, Haeckelidae, Beroida, Thalassocalycida, Dryodoridae(with low support values), Lobata and Cestida, could be very crucial the reconstruction of ancestral characters for the understanding of the plasticity of the characteristic inside this group. This could only achieved by setting a good outgroup for this group.
We strongly recommend for further studies, the identification and posterior description for undescribed species. Also an enrichment of the aligment matrix produced by this study, through sequencing crucial markers such as 18s, ITS1 and NFP, which played an important role in the reconstruction of the phylogeny presented in this study.
Also, we recommend for further studies an extensive sampling of groups like Pleurobrachidae, in an attempt to collapse the long branches presented in previous studies3 and the present study, genus as Tinerfe, which present morphological similarities with families as Haeckeliidae1. More sampling outside groups such as Lobata would allow an improvement for further studies.
The proportion of invariant sites, plays an important role in the analysis of Ribosomal data. During the analysis for this paper, we noticed that the absence of this feature in the analysis in RAxML, forms a clade composed by Beroe and Pleurobrachidae as sister taxa of Lobata, Cestida and Thalassocalycida, this clade was of course with an extremely low bootstrap value; the presence of this feature presents Pleurobrachidae as the sister taxa of Beroe, Lobata, Cestida and Thalassocalycida (Presented in suplementary data). So the absence or presence of this feature during the analysis should be relevant.
The raw data used for this project are available in Zenodo, DOI 10.5281/zenodo.838689 (Arteaga-Figueroa et al., 2016).
LAAF conceived the study, performed the sequence compilation and literature revision. LAAF, VSB and NDFS carried out the phylogenetic reconstructions and analysed the results. All authors were involved in writing the manuscript and have agreed to its final content.
We specially thank Sergio Pulido-Tamayo for stimulating discussions and critical review of the manuscript, Juan F. Díaz-Nieto, Javier C. Alvarez and Diana Rincón T. for their guidance and valuable comments. We also want to thank Lizette I. Quan-Young and Steve Haddock for providing useful bibliography and sequences for this analysis, respectively. Also, we thank to Derrick Zwickl for the comments about model configuration on GARLI.
Supplementary File 1. Rooted tree for the combined dataset (protein + ribosomal DNA) reconstructed by BI using MrBayes. The tree was rooted by midpoint root method and support values (posterior probabilities) are shown on tree nodes.
Click here to access the data.
Supplementary File 2. Rooted tree for the combined dataset (protein + ribosomal DNA) reconstructed by ML using RAxML. The tree was rooted by midpoint root method and support values (bootstrap values) are shown on tree nodes.
Click here to access the data.
Supplementary File 3. Rooted tree for the combined dataset (protein + ribosomal DNA) reconstructed by ML using GARLI. The tree was rooted by midpoint root method and support values (bootstrap values) are shown on tree nodes.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: I am also actively studying the evolution of Ctenophora but I can honestly say that this does not impact my view on the present work.
References
1. Simion P, Bekkouche N, Jager M, Quéinnec E, et al.: Exploring the potential of small RNA subunit and ITS sequences for resolving phylogenetic relationships within the phylum Ctenophora.Zoology (Jena). 2015; 118 (2): 102-14 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
References
1. Simion P, Bekkouche N, Jager M, Quéinnec E, et al.: Exploring the potential of small RNA subunit and ITS sequences for resolving phylogenetic relationships within the phylum Ctenophora.Zoology (Jena). 2015; 118 (2): 102-14 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
|
Version 2 (revision) 21 Aug 17 |
read | |||
|
Version 1 20 Dec 16 |
read | read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
While I am always glad to see new studies on ctenophore phylogeny, I am very surprised that you did not cite Simion et al. 2014 (of which ... Continue reading Dear authors,
While I am always glad to see new studies on ctenophore phylogeny, I am very surprised that you did not cite Simion et al. 2014 (of which I am the first author) for two reasons :
- You used all the data sequenced in that study.
- Both study are very similar in topic and design, and should therefore be compared.
Please find a link to the study : http://www.sciencedirect.com/science/article/pii/S0944200614000816Sincerely,
Paul Simion
While I am always glad to see new studies on ctenophore phylogeny, I am very surprised that you did not cite Simion et al. 2014 (of which I am the first author) for two reasons :
- You used all the data sequenced in that study.
- Both study are very similar in topic and design, and should therefore be compared.
Please find a link to the study : http://www.sciencedirect.com/science/article/pii/S0944200614000816Sincerely,
Paul Simion