Keywords
graph mining, protein-protein interaction, pseudo-absence, semi-supervised learning, support vector machine
This article is included in the Bioconductor gateway.
This article is included in the RPackage gateway.
This article is included in the Interactive Figures collection.
This article is included in the Machine learning: life sciences collection.
Interactions between proteins occur in many, if not most, biological processes. This fact has motivated the development of a variety of experimental methods for the identification of protein-protein interactions (PPIs) networks. Leveraging PPI data available in the STRING database, we use a network-based statistical learning method to infer the putative functions of proteins from the known functions of its neighboring proteins on a PPI network. This package identifies such proteins often involved in the same or similar biological functions. The package is freely available at the Bioconductor web site (http://bioconductor.org/packages/PPInfer/).
graph mining, protein-protein interaction, pseudo-absence, semi-supervised learning, support vector machine
Detailed mathematical description of the support vector machine algorithm with a graph kernel is given in the Methods section. In Figure 1 and 2, log2 transformation of p-values from the enrichment analysis is used in the x-axis. Figure 4 is updated. In Figure 6, the number of interactions we obtained is compared to those between a random set of proteins and the proteins of the RAS pathway by using bootstrapping. Furthermore, we have improved the clarity of the text in several places, according to reviewer's suggestions such as acronyms.
See the authors' detailed response to the review by Helen V. Cook
See the authors' detailed response to the review by Cathy H. Wu and Karen E. Ross
The function of many proteins remains unknown. This is a big challenge in functional genomics. As proteins consisting of the same protein complex are often involved in the same cellular process, the pattern of protein-protein interactions (PPIs) can give information regarding protein function. Thus PPI databases can be useful to predict protein function, complementing conventional approaches based on protein sequence analyses.
Many databases store information on protein interactions and complexes. For example, the Biological General Repository for Interaction Datasets (BioGRID) includes over 500,000 manually annotated interactions1. The STRING database aims to provide a critical assessment and integration of protein-protein interactions, including direct (physical) as well as indirect (functional) associations. The basic interaction unit in STRING is the functional association, i.e. a specific and productive functional relationship between two proteins2.
Once the PPI has been obtained experimentally, there are numerous methods to analyze the network. A neighbor counting method for protein function prediction was developed3,4. The theory of Markov random fields was used to infer functions using PPI data and the functional annotations of its interacting partners5. Many algorithms integrate multiple sources of data to infer functions6–8. We propose a method that can infer the putative functions of proteins by solving classification problem and thus identifying closely connected proteins known to be involved in a certain process.
We use the kernel method for the graph as a similarity measure between proteins, instead of using the first or second level neighbors, and thus our proposed method provides scores or distances derived from a graph kernel. The main advantage of the proposed method is that the neighbors of a set of proteins can be ranked in terms of scores. Generally, we need two or more classes for classification problem. Although we only know the target, we want to apply semi-supervised learning techniques to the PPI through this package. Thus, the main idea of this method is how to find another class so that two classes can be used for classification problem. Eventually, we can classify proteins to identify such closely related proteins by using this package.
Finally, functional enrichment analyses such as over-representation analysis (ORA) and gene set enrichment analysis (GSEA) are incorporated to predict the protein function from the closely related proteins. Although various functional annotations could be used to categorize genes, gene ontology (GO) is one of the most popular function categorization. Kyoto Encyclopedia of Genes and Genomes (KEGG) is commonly used for categorization in pathway analysis. Also, they provide annotations of diverse organisms.
The support vector machine (SVM) is one of the most widely used methods for classification9. Suppose we have a dataset in the real space and that each point in our dataset has a corresponding class label. A SVM is involved in a convex optimization problem to separate data points in the dataset according to their class, by maximizing distance between class and minimizing a penalty for misclassification for each class, at the same time. Unfortunately, the graph data is not in the real space. Cover’s theorem10 provides the useful idea behind a nonlinear SVM, which is to find an optimal separating hyperplane in the high-dimensional feature space mapped by using a suitable kernel function, just as we did for the linear SVM in the original space.
Graph (network) data is ubiquitous and graph mining tries to extract novel and insightful information from data. Graph kernels are defined in the form of kernel matrices, based on the normalized Laplacian matrix for a graph. The best-known kernel in a graph is the diffusion kernel11. The motivation is that it is often easier to describe the local neighborhood than to describe the structure of the whole space12. Another method is called a regularized Laplacian matrix13 and is widely used in areas such as spectral graph theory, where properties of graphs are studied in terms of their eigenvalues and vectors of adjacency matrices14. Broadly speaking, kernels can be thought of as functions that produce similarity matrices15. In the package, we choose only the regularized Laplacian matrix as a graph kernel for the PPI. The kernel K is the symmetric matrix which is given by
where K is the N × N matrix, I is an identity matrix, L is the normalized Laplacian matrix, and γ is an appropriate decay constant.In many biological problems, datasets are often compounded by imbalanced class distribution, known as the imbalanced data problem, in which the size of one class is significantly larger than that of the other class. Many classification algorithms such as a SVM are sensitive to data with an imbalanced class problem, leading to a suboptimal classification. It is desirable to compensate the imbalance effect in model training for more accurate classification. A possible solution to this problem is to use the one-class SVM (OCSVM) by learning from the target class only16. In one-class classification, it is assumed that only information of one of the classes, the target class, is available, and no information is available from the other class, known as the background. The OCSVM can be solely applied because we have only one class, the target. However, it is known that one-class classifiers seldom outperform two-class classifiers when the data from two classes are available17.
In the SVM, the training data set contains m observations x1, …, xm, with corresponding target values y1, …, ym, where yi ∈ −1, 1. Consider the linear classification, wT x + b, w, x ∈ ℝn, and b ∈ ℝ. The distance between the two support lines, or margin, is 2/||w||. Thus maximizing the margin is equivalent to minimizing ||w||/2. The optimization problem is defined as
Here, ξi are slack variables that allow each observation to be on the wrong side of the margin or the hyperplane, while adding a penalization term to the minimization problem. In the SVM, the cost for the misclassification error is controlled by the margin parameter C. For a large value of C, misclassification is suppressed, while for a small value of C, misclassification is allowed for observations that are away from the gathered data18. Consider the dual problem. For the dual variable α ∈ ℝm, we have
In the feature space, the linear decision function is given by wT ϕ(x) + b, using a nonlinear vector function ϕ. Using the kernel K(xi, xj) = ϕT (xi)ϕ(xj), the dual problem in the feature space is
The decision function is given by
for a new observation z. Then the new observation is classified according to the sign of f (z).Unlike the SVM, the one-class SVM is based on the hyperplane approach19. The hypersphere has its center a and radius R. When constructing the hypersphere, its volume should be minimized to tightly encompass observations xi, i = 1, …, m of the target class. Then we have
Here, ξi are slack variables to allow data to lie outside of the hypersphere so as to control the trade-off between the volume and the errors by using the regularization parameter ν between 0 and 1. Its dual problem is
with the dual variable β ∈ ℝm. Given a new observation z, the decision function is given by
where for any support vectors xs. If the value of the decision function is greater than zero, it is classified as a target, otherwise an outlier17.The strategy of this package is to make use of the OCSVM and classical SVM, sequentially. The regularized Laplacian matrix for the whole graph is used as input for them while only nodes with the class label are used for training. Unlabeled nodes are scored as output. First, we apply the OCSVM by training a one-class classifier using the data from the known class only. Let n be the number of proteins in the target class. Also, let I1 be the index sets of rows or columns of the matrix K for the target class. Thus we have
where K* is the n × n matrix which is equal to Kp,q for p, q ∈ I1. This model is used to identify distantly related proteins among remaining N − n proteins in the background by using the matrix Kp,q for p ∉ I1 and q ∈ I1. Indeed, we do not know whether or not each of proteins in the background interacts with the proteins of the target. Thus, it does not always imply that proteins in the background do not interact with the target. In fact, their associations with the target are not observed to date yet. Proteins with zero similarity with the target class are extracted. Then they are potentially defined as the other class by pseudo-absence selection methods20 from spatial statistics. The target class can be seen as real presence data. The main idea of the proposed method is to adopt the pseudo-absence class. For the data to be balanced, assume that two classes contain the same number of proteins. Next, by the classical SVM, these two classes are used to identify closely related proteins whose scores ranging from -1 to 1 are close to 1 among remaining N − 2n proteins. Let I2 be the index sets of rows or columns of the matrix K for these two classes. The corresponding optimization problem is given by where K** is the 2n × 2n matrix which is equal to Kp,q for p, q ∈ I2. The matrix Kp,q for p ∉ I2 and q ∈ I2 is used for scoring. Cross-validation can be used to prevent overfitting and evaluate the above procedure.Semi-supervised learning can be applied to make use of large unlabeled data and small labeled data. Some of these methods directly try to label the unlabeled data. Eventually, those found by this procedure can be functionally linked to the target proteins. This is usually based on the assumption that unannotated proteins have similar functions as their interacting proteins.
We need a list of proteins and the kernel matrix to infer functionally related proteins. With the STRING database for mouse, it is supposed that the target is the set of proteins in the RAS signaling pathway from KEGG pathway (http://www.genome.jp/kegg-bin/show_pathway?mmu04014).
# install necessary packages
source("https://bioconductor.org/biocLite.R")
biocLite(c("PPInfer", "limma", "org.Mm.eg.db", "GO.db")
install.packages("plotly")
library(PPInfer)# download the kernel matrix from https://zenodo.org/record/1066236
download.file("https://zenodo.org/record/1066236/files/K10090.rds", "K10090.rds", mode = "wb")
K.10090 <- readRDS("K10090.rds")# remove prefix
rownames(K.10090) <- sub(".*\\.", "", rownames(K.10090))# load target
library(limma)
kegg.mmu <- getGeneKEGGLinks(species.KEGG = "mmu")
index <- which(kegg.mmu[,2] == "path:mmu04014")
path.04014 <- kegg.mmu[index,1]# infer functionally related proteins
path.04014.infer <- ppi.infer.mouse(path.04014, K.10090, input = "entrezgene",
output = "entrezgene", nrow(K.10090))
genes <- path.04014.infer$top# load gene sets for gene ontology
library(org.Mm.eg.db)
library(GO.db)
xx <- sapply(as.list(org.Mm.egGO2EG), unique)
names(xx) <- AnnotationDbi::select(GO.db, names(xx), "TERM")[,2]# ORA with top 100 proteins
resORA <- ORA(xx, genes[1:100])
head(resORA)
p <- ORA.barplot(resORA, category = "Category", count = "Count", size = "Size",
pvalue = "pvalue", sort = "pvalue", p.adjust.methods = "fdr", numChar = 60,
top = 75) + theme(text = element_text(size = 8), plot.margin = margin(10, 10, 10, 20))# interactive figure
library(plotly)
config(ggplotly(p), showLink = TRUE)The receptor tyrosine kinases (RTKs) bind to a ligand at the cell surface. Then its cytoplasmic domain undergoes conformational change that forms dimerization, resulting in transphophotyrosine. The SH2 (Src homology 2) domain recognizes this phosphotyrosine. The GRB2 (growth factor receptor bound protein 2) protein containing SH2 binds these receptors and recruits SOS1 (SOS Ras/Rac guanine nucleotide exchange factor 1) which is the RAS-GEF inducing RAS to release its GDP and bind a GTP instead. The RAS protein is activated by this guanine nucleotide exchange factor because it has low intrinsic GTPase activity. GTP-activated RAS can activate PI3K (phosphatidylinositol 3-kinase). Once PIP3 (phosphatidylinositol (3,4,5)-triphosphate) is formed by PI3K, an AKT/PKB (Protein kinase B) kinase can become tethered via its PH (pleckstrin homology) domain. When activated, AKT/PKB proceeds to phosphorylate a series of protein substrates, leading to aiding survival by reducing the possibility of an apoptotic suicide program. For this reason, several GO terms about the RTK and PI3K pathways are shown in Figure 1. We can find cell migration due to the integrin and FAK (focal adhesion kinase). MAP (mitogen-activated protein) kinase activity, which is a downstream of RAS signaling and causes cell proliferation, is statistically significant. If a cell has lost anchorage to the extracellular matrix (ECM), it may enter anoikis, a form of the apoptotic cell suicide program. It is known, but poorly understood, that oncoproteins such as SRC and RAS have the ability to mislead a tumorigenic cell into thinking that it has attachment to the ECM in case none may exist at all. We can see the category for negative regulation of anoikis.
# GSEA
index <- !is.na(path.04014.infer$score)
genes <- path.04014.infer$top[index]
scores <- path.04014.infer$score[index]
scaled.scores <- as.numeric(scale(scores))
names(scaled.scores) <- genes
set.seed(1)
resGSEA <- fgsea(xx, scaled.scores, nperm = 1000)
head(resGSEA)
p <- GSEA.barplot(resGSEA, category = "pathway", score = "NES", pvalue = "pval",
sort = "NES", decreasing = TRUE, numChar = 60, top = 75) +
theme(text = element_text(size = 8), plot.margin = margin(10, 10, 10, 20))# interactive figure
config(ggplotly(p), showLink = TRUE)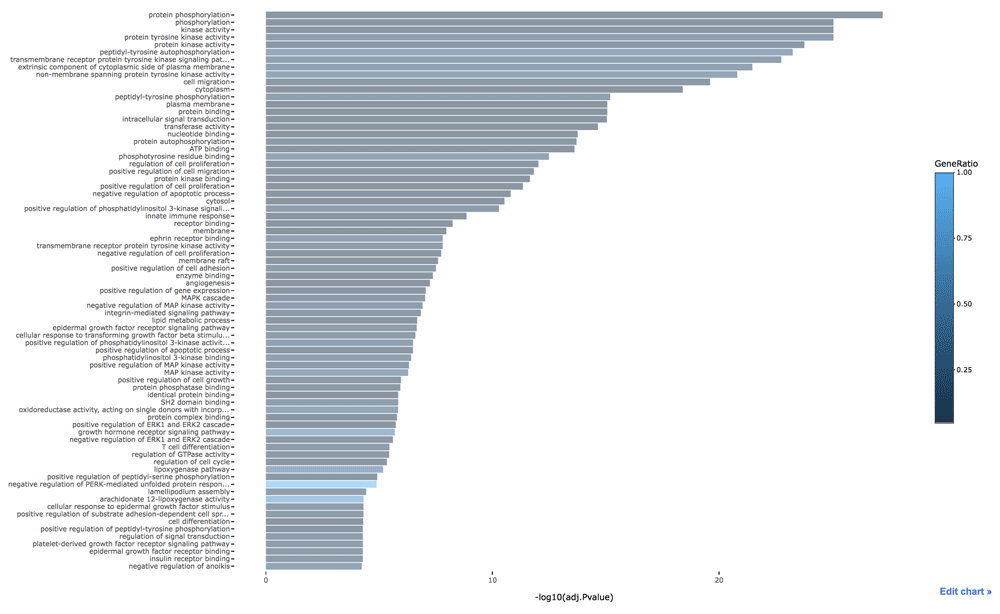
The online version of this figure is interactive.
In GSEA, the gene list is sorted by the standardized score showing how much they are functionally close to the target. Genes with higher scores are functionally more related to the target. The positive values of the enrichment score indicate enrichment at the top of the ranked gene list. Thus proteins that closely related to the target are enriched in categories with high enrichment scores. In other words, these proteins are depleted in categories with low enrichment scores. We can see the RTK, MAPK and WNT signaling pathways in Figure 2. One possible reason is that AKT can activate β-catenin both indirectly and indirectly.
# gene sets for GO and KEGG pathway
library(KEGG.db)
pathway.id <- unique(kegg.mmu[,2])
yy <- split(kegg.mmu[,1], list(kegg.mmu$PathwayID))
library(Category)
names(yy) <- getPathNames(sub("[[:alpha:]]+....", "", pathway.id))
yy[which(names(yy) == "NA")] <- NULL# remove duplicate categories
yy[intersect(names(xx), names(yy))] <- NULL# GSEA
set.seed(1)
GSEAsummary <- fgsea(c(xx, yy), scaled.scores, nperm = 1000)
index <- which(GSEAsummary[,1] == names(yy[1]))
groups <- 0
groups[1:(index-1)] <- "GO"
groups[index:nrow(GSEAsummary)] <- "KEGG"
index <- match(data.frame(GSEAsummary[,1])[,1], names(c(xx, yy)))# network visualization
g <- enrich.net(GSEAsummary, c(xx, yy)[index], node.id = "pathway", numChar = 100,
pvalue = "pval", edge.cutoff = 0.2, pvalue.cutoff = 0.05, degree.cutoff = 1,
n = 200, group = groups, vertex.label.cex = 0.6, vertex.label.color = "black")# interactive figure
vs <- V(g)
es <- as.data.frame(get.edgelist(g))
Nv <- length(vs)
Ne <- length(es[1]$V1)# create nodes
L <- layout.kamada.kawai(g)
Xn <- L[,1]
Yn <- L[,2]
group <- ifelse(V(g)$shape == "circle", "GO", "KEGG")
network <- plot_ly(x = ~Xn, y = ~Yn, type = "scatter", mode = "markers",
marker = list(color = V(g)$color, size = V(g)$size*2,
symbol= ~V(g)$shape, line = list(color = "gray", width = 2)),
hoverinfo = "text", text = ~paste("</br>", group, "</br>", names(vs))) %>%
add_annotations( x = ~Xn, y = ~Yn, text = names(vs), showarrow = FALSE,
font = list(color = "gray", size = 10))# create edges
edge_shapes <- list()
for(i in 1:Ne)
{
v0 <- es[i,]$V1
v1 <- es[i,]$V2
index0 <- match(v0, names(V(g)))
index1 <- match(v1, names(V(g)))
edge_shape <- list(type = "line", line = list(color = "#030303",
width = E(g)$width[i]), x0 = Xn[index0], y0 = Yn[index0],
x1 = Xn[index1], y1 = Yn[index1])
edge_shapes[[i]] <- edge_shape
}# create network
axis <- list(title = "", showgrid = FALSE, showticklabels = FALSE, zeroline = FALSE)
h <- layout(network, title = "Enrichment Network", shapes = edge_shapes,
xaxis = axis, yaxis = axis)
config(h, showLink = TRUE)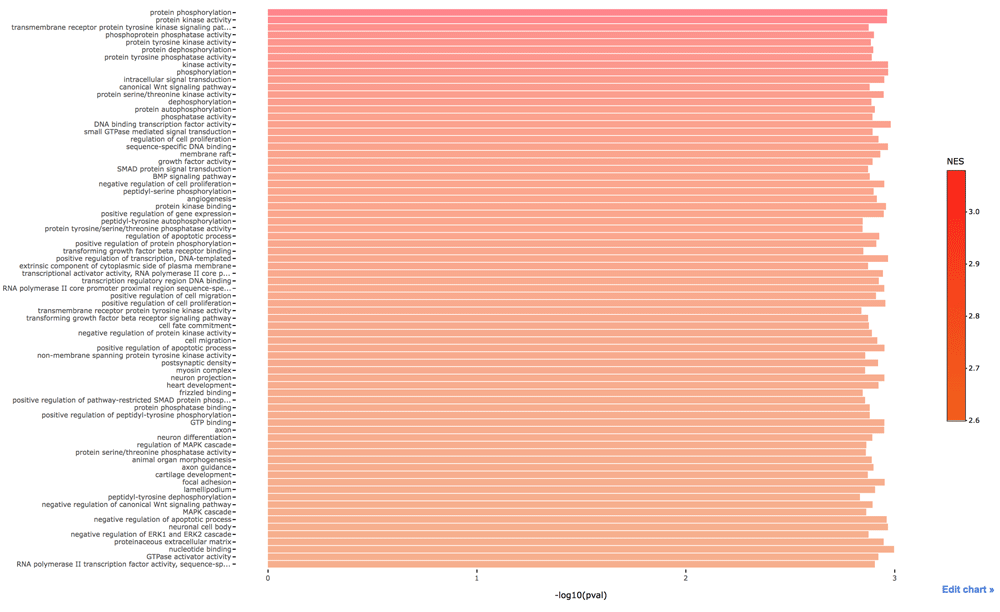
The online version of this figure is interactive.
In Figure 3, the network visualization of the functional enrichment analysis is displayed to minimize the effect of redundancy of gene sets and help in interpretation of enrichment analysis. Nodes indicate gene sets defined by GO terms or KEGG pathways. The connection between two nodes depends on the proportion of overlapping genes of corresponding two gene sets21. The size of nodes is proportional to the number of genes in their categories. The more significant categories are, the less transparent their nodes are. This network may be useful to produce more concise results of the functional enrichment. For GO terms, the two largest subnetworks are involved in transcription and signal transduction. We can see that RAS is related to tumorigenesis via the PI3K-AKT pathway from KEGG.
# top 20 proteins and 30 KEGG pathways
top.genes <- genes[1:20]
filtered.yy <- lapply(yy, intersect, top.genes)
filtered.yy <- filtered.yy[order(lengths(filtered.yy), decreasing = TRUE)[1:30]]
# matrix for heatmap
f <- function(x)
{
index <- match(top.genes, rbind(x))
rbind(x)[index]
}
mat <- ifelse(!is.na(t(sapply(filtered.yy, f))) == TRUE, 1, 0)
# find gene symbol
ensembl <- useMart("ensembl")
mouse.ensembl <- useDataset("mmusculus_gene_ensembl", mart = ensembl)
gene.name <- getBM(attributes = c("mgi_symbol", "entrezgene"), filters = "entrezgene",
values = top.genes, mart = mouse.ensembl)
colnames(mat) <- gene.name[match(top.genes, gene.name[,2]), 1]
# heatmap
library(reshape2)
melt.mat <- melt(mat[c(nrow(mat):1),])
colnames(melt.mat) <- c("pathway", "gene", "value")
p <- ggplot(melt.mat, aes(gene, pathway)) +
geom_tile(aes(fill = value), colour = "white") +
scale_fill_gradient(low = "white", high = "black") +
theme(legend.position = "none", axis.title.y = element_blank(),
axis.text.x = element_text(angle = 90, hjust = 1))
# interactive figure
config(ggplotly(p), showLink = TRUE)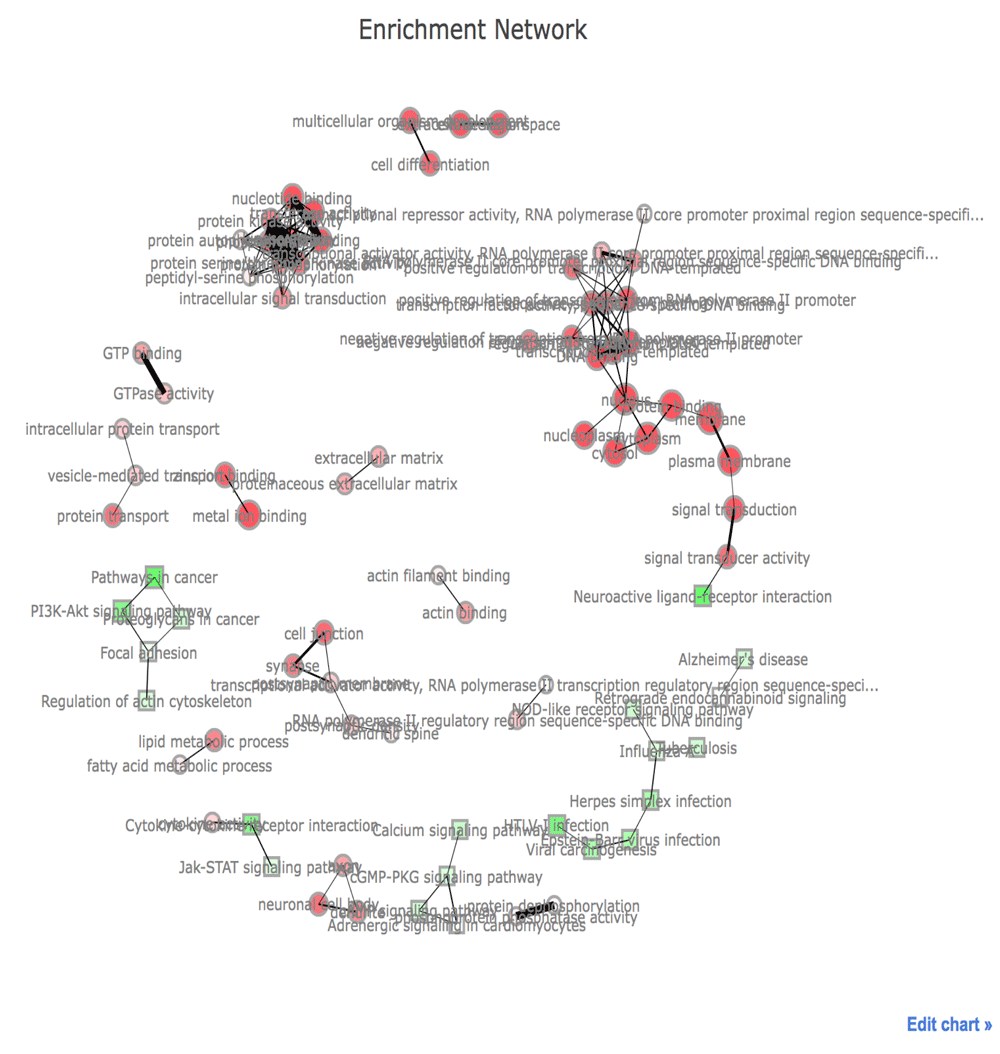
The online version of this figure is interactive.
The binary heatmap for the relationship between top 20 proteins and 30 pathways is shown in Figure 4. The functionally related proteins mainly consist of ErbB receptors and adaptors. Also, they have PI3K and RAF proteins that can be activated by the RAS protein. Pathways containing these proteins are related to known biological functions. However, some of top proteins do not belong to these pathways.
# STRING
string_db <- STRINGdb$new(version = "10", species = 10090, score_threshold = 700)
target <- data.frame(path.04014)
top <- data.frame(top.genes)
names(top) <- "gene.id"# mapping
string_db.map.target <- string_db$map(target, "path.04014", removeUnmappedRows = TRUE)
string_db.map.top <- string_db$map(top, "gene.id", removeUnmappedRows = TRUE)
target <- string_db.map.target$STRING_id
top.proteins <- string_db.map.top$STRING_id# PPI visualization
payload_id <- string_db$post_payload(c(target, top.proteins),
colors = c(rep("#00ff00", length(target)), rep("#ff0000", length(top.proteins))))
string_db$plot_network(c(target, top.proteins), payload_id = payload_id)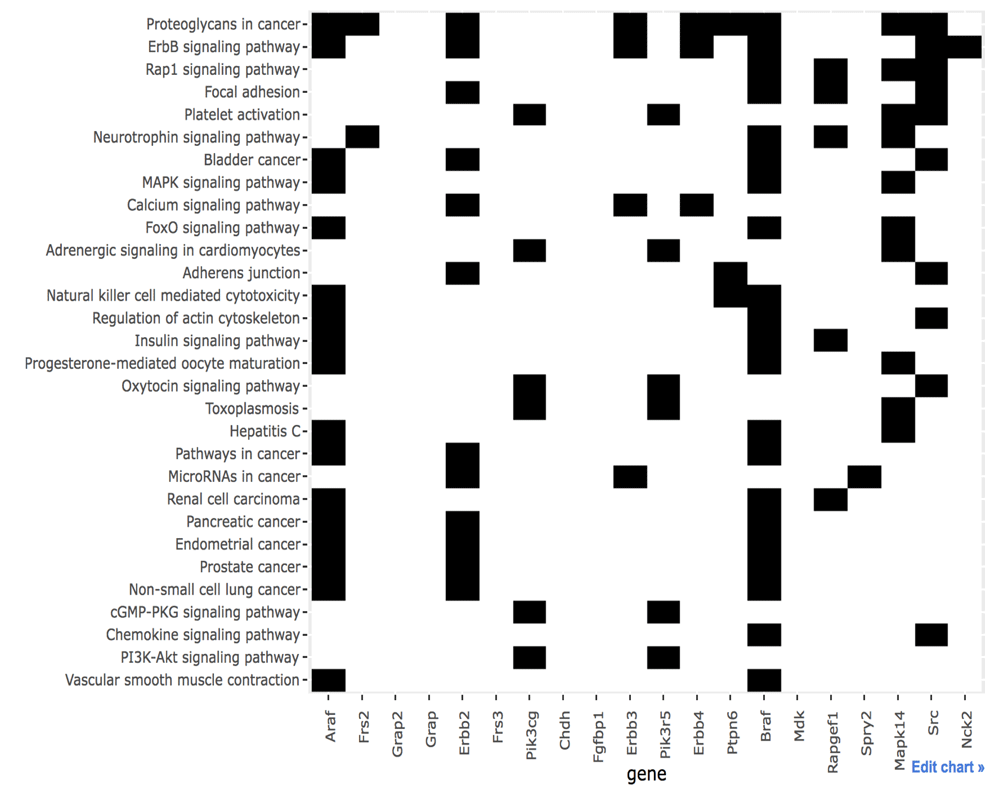
The online version of this figure is interactive.
In Figure 5, the network for high-confidence protein interactions of the RAS pathway and top 20 proteins is drawn by STRING. As expected, top 20 proteins are connected to the target proteins. We can find several types of interactions from the legend of STRING database, but not shown here. For example, interactions between FGFBP1 and the RAS pathway are experimentally determined. On the other hand, interactions with CHDH come from curated databases. CHDH regulated by estrogen in breast cancer is connected to the PLA2 family of the RAS pathway. Also, it is known that PLA2 is phosphorylated by MAPK. Thus the association between CHDH and the RAS pathway is reasonable. Therefore, this approach may be helpful to explain how cytoplasmic mitogenic signaling cascades are activated by estrogen and understand the role of estrogen in breast cancer.
# adjacency matrix
string_db.graph <- string_db$get_graph()
string_db.adjacency <- as_adjacency_matrix(string_db.graph, type = "both")
rownames(string_db.adjacency) <- sub(".*\\.", "", rownames(string_db.adjacency))
colnames(string_db.adjacency) <- sub(".*\\.", "", colnames(string_db.adjacency))# proteins of RAS pathway
path.04014.protein <- getBM(attributes = "ensembl_peptide_id", filters = "entrezgene",
values = path.04014, mart = mouse.ensembl)[,1]
path.04014.protein <- intersect(path.04014.protein, rownames(string_db.adjacency))# first level neighbors of a set of proteins of RAS pathway
string_db.adjacency.path.04014 <- string_db.adjacency[path.04014.protein,]
index <- which(colSums(as.matrix(string_db.adjacency.path.04014)) > 0)
first.nb.protein <- colnames(string_db.adjacency.path.04014)[index]
first.nb.protein <- setdiff(first.nb.protein, path.04014.protein)# first level neighbors of a set of genes of RAS pathway
first.nb.gene <- getBM(attributes = "entrezgene", filters = "ensembl_peptide_id",
values = first.nb.protein, mart = mouse.ensembl)[,1]
first.nb.gene <- intersect(first.nb.gene, genes)# bootstrap
library(boot)
num.overlap <- function(x, index)
{
length(intersect(x[index][1:20], first.nb.gene))
}# observed value
length(intersect(top.genes, first.nb.gene))# resampling
set.seed(1)
(boot.obj <- boot(genes, num.overlap, R = 10000))
boot.num.overlap <- data.frame(boot.obj$t)
colnames(boot.num.overlap) <- "num.overlap.gene"
p <- ggplot(boot.num.overlap, aes(num.overlap.gene)) +
geom_histogram(stat = "bin", binwidth = 0.5) +
labs(x = "Number of overlapping genes", y = "Count") +
scale_x_continuous(breaks = pretty(boot.num.overlap$num.overlap.gene,
n = length(unique(boot.num.overlap$num.overlap.gene)))) +
theme_bw() +
theme(axis.line = element_line(colour = "black"),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.border = element_blank(),
panel.background = element_blank(),
plot.margin = margin(0, 0, 10, 20))# interactive figure
config(ggplotly(p), showLink = TRUE)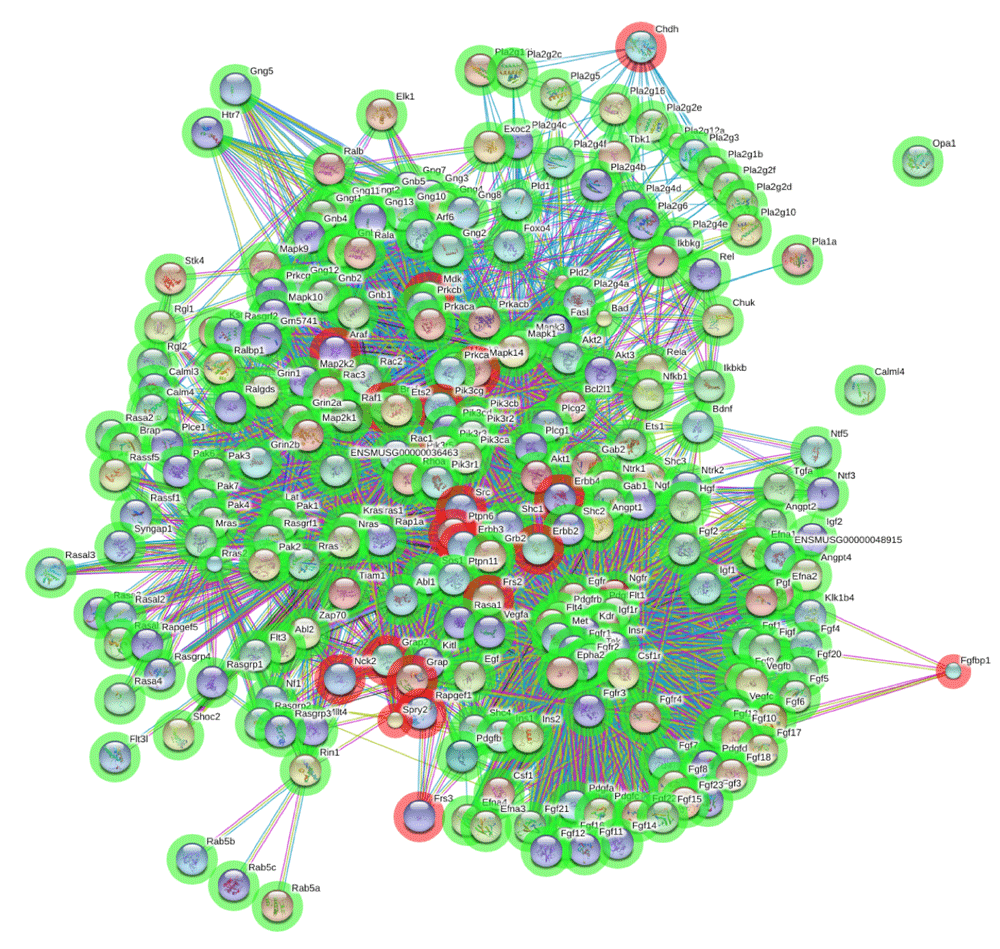
We observe that top 20 proteins are connected to the proteins of the RAS pathway. Similarly, the corresponding genes of the top 20 proteins are connected to those of the target proteins. The first level neighbors of the RAS pathway can be assumed as genes that are connected to the pathway but not chosen by the algorithm. Consider the number of overlapping genes between the first level neighbors and randomly selected 20 genes. Thus, the overlapping genes are randomly selected genes connected to the RAS pathway. The bootstrap method can be used to estimate the distribution of the number of overlapping genes. In Figure 6, the statistic ranges from 0 to 12, based on 10,000 bootstrap replicates. We can conclude that the number of connected genes obtained from the original data is significantly greater than what could be obtained by chance. Therefore, this algorithm seems to be good enough to infer functionally related proteins.
The proposed method is highly dependent on protein interaction networks. There are many databases of protein interactions with millions of predicted protein interactions as well we interactions reported in thousands of publications. Therefore, the choice of data can be critical. Also, the one-class support vector machine can be sensitive to changes in parameters. Although the classical support vector machine has good performance, overall performance may be dominated by the OCSVM since the OCSVM is followed by the SVM. However, it would be worthwhile to provide potential to infer the putative functions of proteins from PPI by complementing conventional methods based on protein sequence analyses.
The PPInfer package is available at: http://bioconductor.org/packages/PPInfer/
Source code is available at: https://github.com/dongminjung/PPInfer
Archived source code as at the time of publication: https://doi.org/doi:10.5281/zenodo.1035128
License: Artistic-2.0
This material is based upon work supported by the National Science Foundation/EPSCoR Grant Number IIA-1355423 and by the State of South Dakota. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
No
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
No
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Is the rationale for developing the new software tool clearly explained?
Partly
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
No
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
No
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
No
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 3 (revision) 12 Mar 18 |
read | |
|
Version 2 (revision) 08 Dec 17 |
read | read |
|
Version 1 07 Nov 17 |
read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)