Keywords
Anopheles gambiae larvae, differentially expressed genes, cadmium, heavy metal tolerance
Anopheles gambiae larvae, differentially expressed genes, cadmium, heavy metal tolerance
In the previous version (check in the materials and methods section-RNA isolation) we had only stated that total RNA that had been extracted was quantified using the micro-spectrophotometer Genequant pro because this was the main equipment that we used for quantification of RNA and we had not recorded the outcome of the quantification.
In the new version, we have included quantification that had also been done for a few samples using the NanoDrop® (Thermo Scientific). The quantification outcome which demonstrates the RNA integrity, has now been recorded in Table 1 for RNA quantification using the micro-spectrophotometer Genequant pro, and in Table 2 for RNA quantification using the NanoDrop® (Thermo Scientific).
RNA is susceptible to degradation due to the presence of RNases in the environment. High quality RNA is required for downstream applications and therefore RNA must be quantified to ensure its integrity. To estimate RNA purity, the ratio of the Absorbance contributed by the RNA to the Absorbance of the contaminants is calculated. The acceptable ratios for purity or typical requirements for A260/A280 ratios are 1.8-2.2.
The extracted RNAs that were considered for the downstream application in Gene fishing Technology were those that met this criterion of purity as stated above.
The added information was in response to the question raised by: Shüné V. Oliver. The other question that had previously been raised by David Essumang, about the generations, I responded to his comment and the information he had requested was already present in the manuscript.
See the authors' detailed response to the review by Shüné V. Oliver
See the authors' detailed response to the review by David Essumang
Heavy metal pollution has become a global environmental problem and severely threatens biological diversity and human health. Our studies on adaptation to heavy metals have documented presence of the mosquitoes in polluted habitats (Mireji et al., 2008) with growing evidence that this adaptation comes at a biological cost to the mosquito (Mireji et al., 2010b). Similar biological costs to adaptations have also been observed elsewhere in Culex pipiens L responses to cadmium, copper, lead and mercury (El-Sheikh et al., 2010). To date, molecular dynamics underpinning heavy metal tolerance in insects have been tied to transcripts and genes associated functionally with immunity (Sorvari et al., 2007) and defense and repair mechanisms such as glutathione transferases and heat shock proteins (Liao & Freedman, 1998; Kim et al., 2000; Stohs et al., 2001). We have previously putatively implicated metallothioneins, alpha-tubulin and cytochrome p450 genes associated with defense, repair and pyrethroid metabolism mechanisms in insects with heavy metal tolerance, using single gene assessment approaches with Anopheles gambiae mosquito larvae (Mireji et al., 2010b; Mireji et al., 2006; Musasia et al., 2013). Here, we have emulated ab initio relatively higher throughput annealing control primer (ACP) transcriptional profiling, to identify:
1) Pathways functionally associated with heavy metal adaptation observed in the field and their associated biological costs (Mireji et al., 2008; Mireji et al., 2010b); and
2) Potential An. gambiae larvae biomarkers that can be applied for assessment of environmental stress or contamination.
Anopheles gambiae s.s mosquitoes that had been collected from the Mbita field station (00.025’S, 34.013’E), Homa Bay County in Kenya were used for the study. The colony was kept in the Animal Rearing and Quarantine Unit (ARQU) at the International Centre of Insect Physiology and Ecology (ICIPE), Nairobi, Kenya. Larval stages of Anopheles gambiae s.s. were selected for tolerance to cadmium heavy metal through chronic exposures of Maximun Acceptable Toxicant Concentration (MATC) that had been empirically determined (Mireji et al., 2010a). Cadmium metal tolerant strains and control (untreated) strains of the mosquito were raised separately and in triplicates. All subsequent generations of the mosquito were subjected to chronic exposures of cadmium metal as described in Mireji et al., (2010a). Standard Operating Procedure (SOP) for the rearing of Anopheles mosquitoes was followed for colony maintenance (Ford & Green, 1972). Cadmium used in our study was applied as Cadmium Chloride (CdCl2) 99.99% pure (Fisher Scientific LLC, Fair Lawn, NJ, U.S.A).
Total RNA was extracted from the third instar larvae of experimental and control An. gambiae populations using Trizol (Invitrogen). Quantification of the extracted RNA was done using the micro-spectrophotometer Genequant pro (Amersham Pharmacia Ltd., Bucks, UK) (Table 1) and a few samples were quantified using the NanoDrop® (Therm Scientific) (Table 2). In addition, DNaseI digestion was carried out to remove any residual DNA that could present in the extracted RNA. Total RNA that was isolated was stored at -80°C.
RNA Quantification was done using the Micro-Spectrometer GeneQuant pro (Amersham Pharmacia Ltd, Bucks, UK).
Quantification was done using the NanoDrop® (Thermo Scientific).
| SAMPLE ID | RNA Concentration | UNIT | A260/A280 |
|---|---|---|---|
| CT1 | 2115.2 | ng/µl | 1.96 |
| CT2 | 3189.2 | ng/µl | 2.0 |
| CD1 | 1824.2 | ng/µl | 2.0 |
| CD2 | 3061.2 | ng/µl | 2.0 |
The total RNA extracted from experimental and control An. gambiae populations were normalized to same concentrations and directly used for the synthesis of first strand complementary DNA (cDNA) using reverse transcriptase (Hwang et al., 2003). Reverse transcription was carried out in a final reaction volume of 20µl containing 2µg of the purified mRNA at 42°C for 1.5 hours. The components of the reaction were: 4µl of 5X reaction buffer (Promega, Madison, WI, U.S.A), 2µl of 10µmol cDNA synthesis dT-ACP 1 primer (5’- CGTGAATGCTGCGACTACGATIIIII(T)18-3’), 5µl dNTPS- 2mM each, 0.5µl RNase inhibitor(40U/µl, Promega) and 1µl Moloney murine leukemia virus reverse transcriptase (200U/µl, Promega). The synthesized first strand cDNA was diluted by adding 80µl ultra-purified water. Storage was at -20°C awaiting PCR procedure.
Annealing control primer based PCR using the GeneFishing™ DEG kit from Seegene, Seoul, South Korea (Kim et al., 2004), was used to determine differentially expressed genes in the heavy metal treated group and the control population.
Synthesis of the second strand cDNA and PCR was carried out in a single tube. The second strand was synthesized in one cycle of first stage PCR at 50°C, in a final reaction volume of 20µl. The components in the reaction tubes included 3–5µl of diluted first strand cDNA, 1µl 10Mm dT-ACP2 reverse primer (5’-CTGTGAATGCTGCGACTACGATIIIII(T)15-3’), 10µl 2x master mix (Seegene, Seoul, South Korea) and 1µl 10µM arbitrary ACP (forward primer).
PCR procedures for the synthesis of the second strand were completed in one cycle, at 94°C for 1 min then 50°C for 3min and 72°C for 1 min.
The second stage of the PCR protocol consisted of 40 cycles at 94°C for 40s, 65°C for 40s, 72°C for 40s and the final extension for 10 min at 72°C. 2% agarose gel electrophoresis with ethidium bromide staining was used for separation of the PCR products.
Differentially expressed bands in the control and cadmium exposed population subjected to the same primer set were excised from the agarose gel using a scapel under Ultra Violet illumination. The gel slices were then purified using the QIAquick® gel extraction kit (QIAGEN, Inc., Valencia, CA), following the instructions from the manufacturer.
Gel-purified PCR products were directly cloned into a pGEMT Easy vector (Invitrogen, Carlsbad, CA, USA), using JM109 competent cells. Colonies were grown at 37°C for 18 hours on Luria broth agar plates, containing ampicillin, X-gal and IPTG for blue/white colony screening. Cloned plasmids were then purified using the GeneJET™ Miniprep kit (Fermentus, Thermo Fisher Scientific Inc.), as per the instructions from the manufacturer.
Sequencing was done with ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA) using M13 primers. The sequences were edited using VecScreen and BioEdit software. Edited sequences were analyzed by searching for similarities in VectorBase against the Anopheles gambiae PEST strain transcripts sequences, AgamP4.6 geneset using the BLASTn search program (Altschul et al., 1990)
We successfully implemented the ACP system to identify differentially expressed genes (DEGs) in larvae chronically exposed to cadmium, as previously demonstrated in blastocyst experiments (Cui et al., 2005; Hwang et al., 2004; Hwang et al., 2005). Our differential banding patterns of the cDNA representation of DEGs is summarized in Figure 1. Fourteen DEGs were identified after chronic exposure of An. gambiae larvae to cadmium heavy metal (Table 3). Most (11) of the differentially expressed genes were induced in cadmium exposed relative to the cadmium un-exposed controls. Our BLAST (REF) results revealed that the cadmium induced transcripts were clustered into metabolism (AGAP008584-RA, AGAP001249-RA and AGAP009563-RA), transport (AGAP012302-RA and AGAP002638-RA) and protein synthesis (AGAP028915-RA, AGAP004750-RA, AGAP028391-RA, AGAP003870-RA, AGAP028907-RA, AGAP028818-RA and AGAP028899-RA) processes.
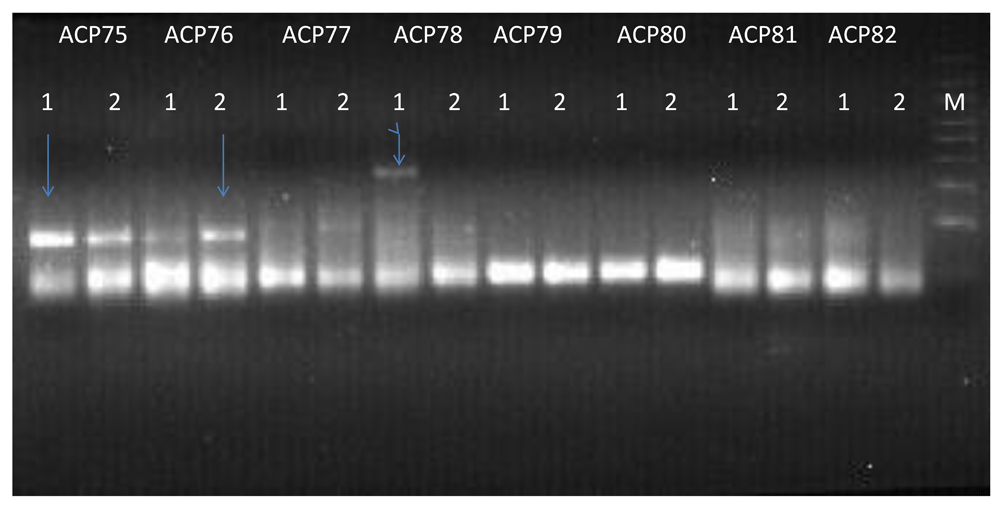
The arrows indicate the DEGs observed using ACP 75, ACP 76 and ACP 78 primer set. Number 1 represents Cadmium population while 2 represents control population. M= 50bp molecular marker. High intensity of a band represents an up-regulation of a particular gene in cadmium or control population.
Sequence data obtained was blasted against Anopheles gambiae PEST strain transcript sequences, AgamP4.6 geneset in May 2017.
Three of the DEGs identified were suppressed in the cadmium exposed larvae and these included AGAP006187-RA, AGAP002262-RA and AGAP003078-RA.
We identified ATP-binding cassette transporters belonging to the superfamily of membrane proteins that are present in all living organisms (Dean & Annilo, 2005). They are normally associated with movement of substrates such as amino acids, peptides, sugars, metals, inorganic ions, lipids, lipopolysaccharides and xenobiotics across biological membranes (Dawson & Locher, 2006; Hollenstein et al., 2007a). The ABC transporters have been shown to affect development, metabolism and insecticide resistance in insects (Borycz et al., 2008; Dow & Davies, 2006; Ricardo & Lehmann, 2009; Vache et al., 2007). The silencing of the ABCH1 gene has been shown to result in the death of larvae and pupae (Guo et al., 2015). Therefore, induction of the ABC transporters may suggest that they are involved in cadmium transport through membranes to reduce toxicity of cadmium metal to the larvae in their environment.
The induction of the eupolytin gene may have a role in the activation of defense molecules. In a study involving the ground beetle Eupolyphaga sinensis, eupolytin-1 gene encoding a protease was shown to be involved in the activation of plasminogen and hydrolysis of fibrinogen (Yang et al., 2011).
Ribosomal genes are involved in protein synthesis and upregulation of the various arrays of ribosomal RNAs in this study, which suggests their role in enhancing the survival of An. gambiae in the heavy metal polluted environment by the transcription and translation of genes which are important in the adaptation of the larvae to xenobiotics.
The nuclear structure referred to as THO complex is usually conserved in all kingdoms, and it has an important role in the packing of pre-mRNA molecules into RNA-protein assemblies which are termed mRNPs (Köhler & Hurt, 2007). A study carried out recently has shown that the THO complex is required for efficient expression of some genes, ensuring genetic stability thereby preventing transcription-associated recombination (Gewartowski et al., 2012). The expression of the THO complex is suggestive of its role in expressing defense genes that enhance survival of larvae in a Cadmium polluted environment.
Suppression of AGAP006187-RA, AGAP002262-RA and AGAP003078-RA transcripts that included G- Proteins couple receptors to adenylyl cyclase stimulation indicated increasing levels of cAMP and a cascade of events that constitute the signal transduction pathway that drive cellular responses. Metallopeptidases are a ubiquitous and diverse group of enzymes containing both endopeptidases and exopeptidases. Although they vary widely at the sequence, structural, and functional levels, they all require a metal ion for catalytic activity (Rawlings & Salvesen, 2013). The suppression of these important genes involved in signal transduction and proteolytic activity would account for the larval mortality rates that are usually observed in larvae raised in the cadmium heavy metal environment.
Our findings shed light on the adaptation of the An. gambiae mosquito to heavy metals by differentially expressing particular genes in response to a toxicant impact. A study to determine differentially expressed genes in cadmium-exposed sebastes schlegeli unraveled genes related to pathogenesis, extrinsic stresses, and catalytic metabolites (Woo & Yum, 2014). Previous studies have indicated that metallothionein and α-tubulin genes that are present in insects can be used as potential biomarkers (Hare, 1992; Klerks & Weis, 1987; Mattingly et al., 2001; Roesijadi, 1994). Metallothionein was assessed through C. quinquefasciatus mosquito larvae for Copper, Cadmium and Zinc aquatic environmental levels (Sarkar et al., 2004). Therefore, the genes identified might be useful in the development of potential biomarkers that can be used to assess the level of environmental pollution of heavy metal origin in An. gambiae mosquitoes.
We were able to identify genes that are differentially expressed due to chronic exposure of An. gambiae larvae to cadmium metal using the ACP-based PCR method. However, application of more sensitive techniques like those used in proteomics would generate more data.
Dataset 1: Sequence data obtained after sequence analysis using the BioEdit software. The sequences were subsequently taken through a BLAST search. The results of the sequence analysis are shown on the manuscript. DOI, 10.5256/f1000research.13062.d187045 (Muturi et al., 2017a).
Dataset 2: Sample of the colony PCR experiment. The gel photo of a colony PCR of 20 samples that was carried out after blue/white colony screening using M13 primers. DOI, 10.5256/f1000research.13062.d187046 (Muturi et al., 2017b).
Funding for this study was provided by the Department of Research and Extension, Egerton University and the DAAD in-country Scholarship.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We hereby wish to acknowledge the following individuals for their contribution to this work:
The Head of the Capacity Building Department at ICIPE, for granting us permission to carry out this work in their Molecular and Biotechnology unit.
The Director of the Research and Extension Department at Egerton University.
The DAAD team for the financial support, which enabled this work to be completed.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
No
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Ecological genomics, transcriptomics, comparative genomics. Molecular ecology of stress adaptation
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Environmental Scientist
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 25 Jun 18 |
read | read | |
|
Version 1 22 Dec 17 |
read | read | |
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Click here to access the data.
Spreadsheet data files may not format correctly if your computer is using different default delimiters (symbols used to separate values into separate cells) - a spreadsheet created in one region is sometimes misinterpreted by computers in other regions. You can change the regional settings on your computer so that the spreadsheet can be interpreted correctly.
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)