Keywords
RNA-seq, Android-based, simulations, mobile application, recommendations, experimental design
RNA-seq, Android-based, simulations, mobile application, recommendations, experimental design
Keeping in view the reviewers’ suggestions, we have made the following changes in the revised version of the manuscript.
See the authors' detailed response to the review by Niranjan Nagarajan
See the authors' detailed response to the review by Daisuke Komura
RNA-seq offers several advantages over low-throughput technologies such as quantitative PCR and annotation-dependent methods such as microarrays. Designing RNA-seq experiments accurately, however, poses challenge to biologists. This is particularly true when prior knowledge on genome or transcriptome of the organism of choice is not available. It is important to determine the number of technical replicates and the number of sequencing reads, and choose the right analytical tool, to estimate subtle differences between expression levels of transcripts.
Web-based tools, Scotty (Busby et al., 2013) and EDDA (Luo et al., 2014), have an established precedence in aiding RNA-seq design. While Scotty relies solely on pilot or prototype data, EDDA relies on either pilot data or a simulate-and-test paradigm to account for variability across experimental conditions. Scotty has a built-in t-test based module, whereas EDDA has been linked to five other DE tools, post mode-normalization of the data. Both can detect DEGs upto 2-fold difference.
In the current manuscript, we describe RNAtor, an Android app with a user-friendly graphical user interface (GUI) that helps biologists design RNA-seq experiments. A mobile application offers a lot more flexibility, ease of navigation, user-friendliness, and offline features compared to a web-based tool, even when the latter can also be accessed or computed on the mobile. RNAtor can be linked to any existing differential expression analysis tool, and can help design experiments to estimate expression differences with as low as 0.8–1.2X fold change. RNAtor’s recommendations are based on an exhaustive combination of discovery with simulated reads for transcriptomes of varying sizes (3 to 100 Mb). These recommendations are subsequently validated with sequenced data from Saccharomyces cerevisiae, while comparing expression profiles of wild-type and mutant strains.
We simulated varying numbers of Illumina-like reads with technical replicates, with fold changes ranging from 1.2–5X between the control and treatment samples, in both directions, on a 3 Mb human chr14 (hg19) transcriptome, using Polyester (Frazee et al., 2015). We detected differentially expressed genes (DEGs) on all the simulations using Tophat v2.1.1-Cufflinks v2.2.1 (Trapnell et al., 2012) based genome-guided workflow followed by differential expression analyses using five tools: Deseq v1.28.0 (Anders & Huber, 2010); Deseq2 v1.16.1 (Love et al., 2014); EdgeR v3.18.1 (Robinson et al., 2010); Cuffdiff-Cufflinks v2.2.1 (Trapnell et al., 2012); and Kallisto v0.43.1 (Bray et al., 2016) and a de novo assembly-based tool, Trinity v2.3.2 (Grabherr et al., 2011) followed by differential expression analyses using Kallisto v0.43.1 (Bray et al., 2016). Thus, Kallisto was used twice; first, with the genome-guided paradigm and second, with de novo assembly using Trinity. In the first scenario, the Tophat-Cufflinks alignments (.bam) were converted to reads (.fastq) to be used with Kallisto along with the 3 Mb transcriptome as the reference. In the second scenario, the de novo assembled transcriptome as the reference along with the simulated reads was used with Kallisto. All differential expression analysis softwares were run with default cut-offs. We studied results from these simulations on the number of DEGs detected reliably and the extent of recovery of those DEGs. Transcript recovery refers to the length the transcript as assembled by Tophat, found to be differentially expressed by EdgeR or CuffDiff or DESeq2, in relation to the actual length as per simulations. It is possible to estimate this parameter only for these three tools, since they offer a handle to the actual transcript IDs. Based on these simulations, we arrived at recommendations on the number of reads, number of replicates, and the tool(s) needed to identify DEGs reliably. We validated these recommendations using simulated reads from larger transcriptomes (10Mb, 30Mb and 100Mb), created by combining transcriptomes from more than one hg19 chromosome, and using a real Sacharomyces cerevisiae dataset (ENA accession: ERP004763) comprising of 48 biological replicates, for two conditions; wild-type (WT) and a snf2 knock-out (KO) mutant (Schurch et al., 2016).
The size of the transcriptome (or genome if the transcriptome size is not known), taken from a user-defined or from a backend database, the number of replicates to use and the fold change of DEGs are user-defined parameters in RNAtor (Figure 1). An RNAtor flowchart highlighting simulation conditions and analytical tools used is provided in Supplementary Figure S1.
RNAtor was evaluated using questions that a biologist would typically ask before starting an experiment, followed by the recommendations provided by RNAtor.
One, 1.5, 6, 10, 14 and 20 million reads are needed for detection of differential expression of DEGs at 5-fold, 4-fold, 3-fold, 2-fold, 1.5-fold and 1.2-fold change, respectively, for a 3Mb transcriptome with 3 technical replicates.
We simulated 0.2–20 million reads for human chromosome 14 (~3Mb) and observed that the numbers of detected DEGs simulated at a given fold change peaked for a certain coverage before plateauing (Figure 2). This observation remained valid for the real data (Figure 3) and the large simulated transcriptomes (10Mb, 30Mb and 100Mb) (Supplementary Figure S2). Increasing the number of sequencing reads increased the sensitivity of detection. The final recommendations from RNAtor correspond to the number of DEGs at its peak, and are therefore, a good compromise between sensitivity and keeping the cost of sequencing low. Changing the number of technical replicates does change the recommendation. For example, with more than three replicates, RNAtor suggests producing fewer reads to obtain the same information (Table 1).

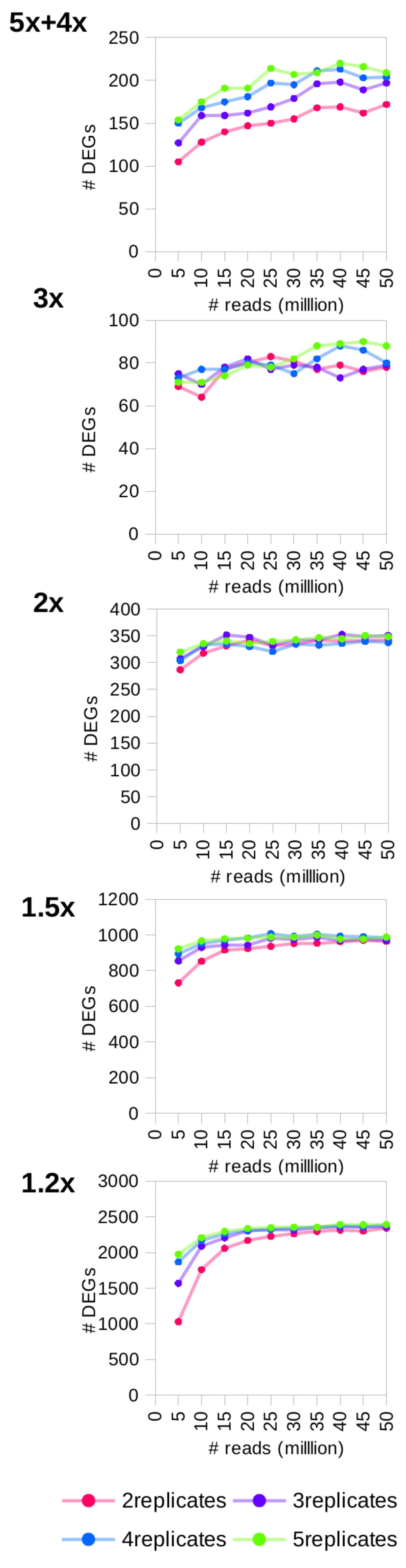
Kallisto detected optimal number of DEGs with the highest sensitivity. Focusing purely on the number of DEGs detected between WT and KO, Kallisto performed best over the other tools tested (Figure 2 and Supplementary Figure 3).
Cuffdiff can be used for high specificity and DeSeq2 and EdgeR, for high transcript recovery. Although Kallisto-Sleuth was fast and produced results with high sensitivity; we observed that this was at the expense of specificity of detection (Supplementary Figure S3). Cuffdiff produced results with high specificity albeit with a loss of sensitivity (Supplementary Figure S3). The transcript recovery was best for EdgeR for shorter (<742 bases) and medium-sized (742–1456 bases) transcripts, and best for CuffDiff for longer transcripts (>1456 bases), among the 3 tools tested (CuffDiff, DeSeq and EdgeR, Supplementary Figure S4).
The assembly-based pipeline yields more DEGs with higher sensitivity and specificity. Using Trinity (Grabherr et al., 2011) as an assembly pipeline along with Kallisto enhanced the number of DEGs detected when compared with the genome-guided Kallisto-Sleuth pipeline (Figure 2). While the sensitivity of Trinity-Kallisto was marginally better, its specificity was visibly better when compared to the Kallisto-Sleuth pipeline (Supplementary Figure S3).
Although some of the challenges with RNA-seq experiments have been addressed previously (Busby et al., 2013; Luo et al., 2014), currently there is no easy-to-use, biologist-friendly mobile phone-based app. Scotty, a previously reported, useful, interactive web-based tool aids RNA-seq experimental design. However, it has a dependence on pilot or prototype data, closely matching the actual experimental conditions (Busby et al., 2013). EDDA, another web-based interactive RNA-seq experimental design aiding tool, offers more flexibility in terms of the use either providing pilot data or using a simulate-and-test paradigm as per the desired experimental conditions (Luo et al., 2014). Both can detect genes or transcripts of only up to 2X fold change in the test condition relative to the control. RNAtor addresses some of these gaps as a user-friendly mobile app. Hhowever, it has certain limitations. For example, it does not take into account the dynamic nature of any transcriptome (where the exact size of transcriptome is not known and cannot simply be derived from the genome size), the throughput of different sequencing instruments, the presence of spliced variants, and the relative abundance of transcript, for e.g. in relation to a control gene or any other gene of interest. We also recognize that the RNAtor v1.0 is based on simple assumptions that can affect the recommendations. Nevertheless, the validation of the recommendations resulting from training on simulated RNA-seq data that has not yet incorporated various biological biases, with real data from Saccharomyces cerevisiae provides strong evidence that our assumptions do not significantly impact RNAtor's guidance to users. That said, there is a prevailing need for a simple tool for biologists, who have simple questions. RNA-seq is not necessarily used to answer complex questions always, but also often as a superior substitute to qPCR. We intend to expand the scope of the tool in its future releases, by introducing biases that mimick various experimental conditions into the simulation phase.
The Android version of RNAtor is available on Google Play Store.
Latest source code: https://github.com/binaypanda/RNAtor.
Archived source code as at the time of publication: https://doi.org/10.5281/zenodo.814905 (Panda, 2017).
License: RNAtor v1.0 is distributed under GNU GPLv3 licence.
Research presented in this article is funded by the Department of Electronics and Information Technology, Government of India (Ref No: 18(4)/2010-E-Infra., 31-03-2010) and Department of IT, BT and ST, Government of Karnataka, India (Ref No: 3451-00-090-2-22).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary Figure S1: RNAtor flowchart highlighting simulation conditions (reads, technical replicates, and fold change of differential expression) and analytical tools used.
Click here to access the data.
Supplementary Figure S2: Number of differentially expressed genes (DEGs) detected for various simulated dataset on 10Mb, 30Mb and 100Mb transcriptomes using the Kallisto-Sleuth pipeline.
Click here to access the data.
Supplementary Figure S3: True/false positive curves for differentially expressed genes (DEGs) recovered under various simulation conditions, created by combining reads (0.1M–20M), technical replicates (2–5) and fold change of differential expression (1.2–5X) by Cuffdiff, Deseq2, EdgeR, Kallisto and Trinity-Kallisto tools.
Click here to access the data.
Supplementary Figure S4: Percentage recovery of transcripts under various simulation conditions, created by combining reads (0.1M–20M), technical replicates (0–5) and folds change of differential expression (1.2–5X) with CuffDiff, DeSeq and EdgeR. The size of the bubble represents the extent of transcript recovery.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: My lab developed the software EDDA (http://edda.gis.a-star.edu.sg/; https://genomebiology.biomedcentral.com/articles/10.1186/s13059-014-0527-7) which has partly overlapping functionality.
Reviewer Expertise: Genomics, Computational Biology
Is the rationale for developing the new software tool clearly explained?
No
Is the description of the software tool technically sound?
No
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
No
Competing Interests: My lab developed the software EDDA (http://edda.gis.a-star.edu.sg/; https://genomebiology.biomedcentral.com/articles/10.1186/s13059-014-0527-7) which has partly overlapping functionality.
Reviewer Expertise: Genomics, Computational Biology
Is the rationale for developing the new software tool clearly explained?
Partly
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 16 Nov 17 |
read | read |
|
Version 1 26 Jun 17 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)