Keywords
HBV/C2, Chronic, Non-recombinant, Bangladesh
HBV/C2, Chronic, Non-recombinant, Bangladesh
The number of cases of chronic liver disease caused by hepatitis B virus (HBV) are increasing markedly1. Globally, more than 2 billion people have been infected by HBV2 and, according to the World Health Organization (WHO) approximately 257 million were living with HBV in 2017. In Bangladesh, the rate of HBV chronicity is 2–6%3, which makes it relatively higher risk than other infectious diseases.
HBV genome comprises a partially double-stranded covalently closed circular DNA that encodes four highly overlapping major open reading frames4. Due to the absence of proof-reading activity, the mutation rate of HBV is high5; hence, recombinant strains are evolving with a common pattern. Most of the HBV cases are chronic, which has a high possibility of causing liver cirrhosis6 and hepatocellular carcinoma7. In Bangladesh, there are no reported reference complete sequence of HBV chronic strain of subgenotype C2. Hence, we isolated the complete genome of a HBV/C2 strain collected from a patient without liver complications, though carrying the virus for a long time.
An HBV-positive plasma sample was collected from a 45-year-old male patient in a tertiary hospital in Dhaka, Bangladesh after obtaining the patient’s written informed consent. The infected patient had chronic liver disease, as determined by ultrasonography. The patient was diagnosed with chronic HBV infection recently, with a high viral load in the plasma. However, the patient was not showing signs of jaundice, though was affected by fever, nausea, vomiting and fatigue. The study was approved by the Research Ethics Committee of National Institute of Biotechnology, Bangladesh (NIBREC2015-01). The patient was not taking any antiviral therapy and was diagnosed 1 month prior to obtainment of the plasma sample. HBV DNA was extracted from the sample using the QIAamp MinElute Virus Spin kit (Qiagen, Germany). The complete HBV genome was amplified by six sets of primer pairs used previously in another study8 using a conventional PCR method. The primer sequences and their annealing temperatures were as follows: set 1, forward- AAGCTCTGCTAGATCCCAGAGT, reverse- AGTTGGCGAGAAAGTGAAAGCCTG, 56°C; set 2, forward- CCTATTGATTGGAAAGTATGTCA, reverse- AACAGACCAATTTATGCCTA, 48°C; set 3, forward- GAGACCACCGTGAACGCCCA, reverse- CCTGAGTGCTGTATGGTGAGG, 56°C; set 4, forward- TTCACCTCTGCCTAATCATC, reverse- ATAGGGGCATTTGGTGGTCT, 52°C; set 5, forward- TCAGGCAACTATTGTGGTTTCA, reverse- GGGTTGAAGTCCCAATCTGGATT, 51°C; set 6, forward- GGGTCACCATATTCTTGGGAA, reverse- CGAGTCTAGACTCTGTGGTA, 51°C. For a mixture of 25 µl reaction volume, 12.5 µl of 2X MasterMix (Thermo Fisher Scientific, USA), 1 µl each of forward and reverse primers (IDT, USA), 9.5 µl of nuclease-free water (Thermo Fisher Scientific, USA) and 2 µl of template DNA were used. The condition of the PCR reaction was 1 cycle at 95°C for 10 min, 35 cycles at 95°C for 1 min, with the aforementioned annealing temperatures for 1 min and 72°C for 1 min, and a final cycle for 10 min at 72°C. Sanger sequencing was performed using the BigDye Terminator version 3.1 cycling sequencing kit (Applied Biosystems, USA) by ABI 3130 Genetic Analyser (SeqGen, CA, USA) and by thermal cycler (Sigma-Aldrich, Germany) using the described annealing temperatures as per manufacturer’s instructions after the purification of PCR products using PureLink PCR Purification Kit (Thermo Fisher Scientific, USA), performed in accordance with the manufacturer’s protocol. Next, the sequenced contigs were assembled using the Seqman tool of DNASTAR Lasergene version 7.29.
The subgenotyping and mutation analysis of the sequenced genome were performed using the HBV Geno2Pheno tool version 2 using the default parameters, comparing against the HBV genotype D consensus sequence. Recombination analysis of the sequence was performed using the NCBI genotyping tool. The complete genome was deposited in the GenBank under the accession number MH220971.
Analysis of the complete genome denotes that the isolate studied here, termed NHB17965, comprises HBV genotype C and subgenotype C2 (HBV/C2) with a GC content of 48.77%. Recombination analysis using the NCBI Genotyping tool showed that NHB17965 is a non-recombinant wild-type HBV isolate (Figure 1).
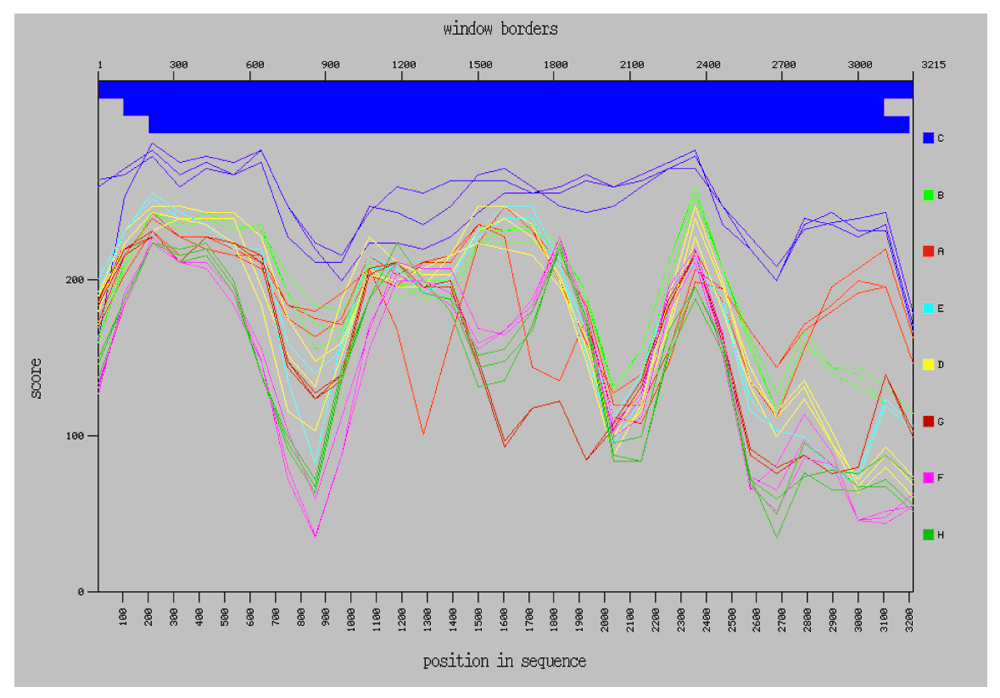
The Simplot diagram was generated using the NCBI Genotyping tool.
Isolate NHB17965 was observed to have amino acid substitutions H9Y, N13H, I91L, P109S, T128N, I269L and V278I in the polymerase domain and S53L, P120T, I126T and S210N in the small hepatitis B surface protein as analysed by HBV Geno2Pheno tool, compared against the HBV genotype D consensus sequence. These substitutions may be the results of regular genomic changes to HBV because of a lack of proof-reading activity of the viral reverse transcriptase, and may not signify any danger. Hence, the isolate NHB17965 could be considered as a reference strain of chronic HBV/C2 infection in Bangladesh.
The findings of this study will help clinicians and scientists to gain substantial knowledge about the current common genomic substitutions of HBV/C2 and to develop treatments against chronic HBV infections.
Genome of the HBV strain isolated in this study, MH220971.
The study was supported by the National Institute of Biotechnology, Ministry of Science and Technology, Bangladesh.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 3 (revision) 10 Sep 18 |
read | ||
|
Version 2 (revision) 13 Aug 18 |
read | read | |
|
Version 1 09 Jul 18 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)