Keywords
sialidases, Influenza A, defence strategy, exogenous expression, α(2,3)-sialylation
This article is included in the Emerging Diseases and Outbreaks gateway.
sialidases, Influenza A, defence strategy, exogenous expression, α(2,3)-sialylation
The global influenza pandemic remains a real threat. Avian influenza viruses H5N1 have killed millions of birds, including domestic poultry, causing enormous financial losses. Several thousands of cases of human infection were recorded and some with a fatal outcome (see World Health Organisation (WHO) assessment of Avian Flu pandemic threat). The risk of a new pandemic of viral infection remains high. Seasonal flu vaccination is used as a traditional way to prevent the infection spread, however it has a lot of limitations. Therefore, it is critical to develop alternative approaches to prevent influenza infection. An alternative way to reduce the infection could be the creation of genetically modified domestic birds. It will decrease the risk of the infection spread, because domestic birds are known to transmit the infection acting as an intermediary between wild ducks to humans (Kim et al., 2009). It is necessary to select gene-candidates for developing the approach.
The best way to prevent the infection is to impede the virus from entering the cell. Influenza virus hemagglutinin (HA) provides attachment to the host cell that leads to fusion between the virion envelope and the host cell membrane (Skehel & Wiley, 2000). Surface sialic acid (SA) residues are host cell epitopes that are recognized by influenza virus A and B (Ito, 2000). SA are a family of nine-carbon acidic monosaccharides that naturally terminate sugar chains attached to the proteins on the cell surface (Varki et al., 2009). Neuraminidase (NA) - the second major surface antigen - is an exoglycosidase, or saildase, that cleaves SA from cell membrane glycolipids and glycoproteins, thus destroying the recognition epitopes on the surface of the host cell for the viral receptor HA. NA activity helps viral particles penetrate through mucous secretions that are rich in sialic acids, to reach the target cells in the airway epithelium (Palese et al., 1974). The role of the enzyme in facilitating release of newly formed viral particles from the infected cell surface and preventing aggregation of the viral particles was experimentally confirmed (Matrosovich et al., 2004; Palese et al., 1974). There are known antiviral agents - oseltamivir and zanamivir - that inhibit NA, blocking the release of virus particles from infected cells (Moscona, 2005; Varghese, 1999). Sialidases have also demonstrated to be effective inhibitors of Influenza virus infection in vitro. It has been shown that cells treated with Vibrio cholerae or Micromonospora viridifaciens bacterial sialidase are resistant to influenza virus infection (Air & Laver, 1995; Bergelson et al., 1982; Griffin et al., 1983; Stray et al., 2000).
We focused on exogenous expression of sialidases as a defense strategy against influenza infection (Figure 1). Here we show the protective effect of exogenous expression of different sialidases. The range of exogenous sialidases has diverse activity and specificity towards sialic acid residues. Tissue-specific expression of sialidases in transgenic poultry might protect domestic birds against Influenza virus.
Viral neuraminidase. Viral neuraminidase from human Influenza A virus was amplified from the plasmid pNAHA (Plasmid # 44169; Addgene, Cambridge, MA, USA) that encodes the neuraminidase gene of Influenza A / Puerto Rico / 8/34, subtype N1. The amplified product was inserted by restriction cloning in the lentiviral transfer plasmid based on a pHAGE backbone (#24526, Addgene) after the CMV promoter. The RFP reporter was cloned in the same reading frame and the resulted construction was pHAGE-CMV-infNA-P2A-RFP (Supplementary Figure 1A).
Sialidases from Salmonella typhimurium and Actinomyces viscosus. Synthesis of the codon-optimized bacterial sialidase from Salmonella typhimurium (Uniprot / P29768, 2-382aa) and the codon-optimized bacterial sialidase from Actinomyces viscosus (uniprot / Q59164, 347-631aa) fused with the transmembrane domain of viral neuraminidase (1 - 75 aa) were done in BioCat. The transmembrane domain was added to provide membrane localisation of the enzymes, since sialidase Salmonella.t. and A. viscosus are cytoplasmic. The codon-optimization was done with the IDT tool with the choice of the genome of Gallus gallus. The genes were ordered in the standard vector pUC57 and were flanked by the restriction sites NheI and SphI for S. typhimurium and XhoI and SphI sites for A. viscosus. The genes were excised from the vector pUC57 using the sites and inserted into the pHAGE backbone after the the CMV promoter. As a result, the plasmids pHAGE-CMV-Sal.t.Sia-P2A-RFP and pHAGE-CMV-Act.v.Sia-P2A-RFP have been cloned (Supplementary Figure 1A).
The genes coding sialidase 3 (membrane sialidase) in G.gallus and H.sapiens are homologous based on the analysis in HomoloGene (NCBI). We selected human membrane sialidase (Gene ID: 10825) for cloning because there is more information about it compared with other orthologs. The coding exons were amplified from human genomic DNA and overlapped. The resulted product was inserted by restriction cloning in the lentiviral transfer plasmid based on the pHAGE backbone (#24526, Addgene) after CMV promoter. The RFP reporter was cloned in the same reading frame and the resulted construction was pHAGE-CMV-hNeu3-P2A-RFP (Supplementary Figure 1A).
Enzymatically inactive neuraminidase was taken as a negative control. Two amino acids at positions 262-263 (EE) of the Influenza A virus strain A/Puerto Rico/8/1934 H1N1 are responsible for the substrate binding. The inactive variant can be obtained by the mutation E262D (Huang et al., 2008). The mutation was made by two overlapping primers: 5’-GCACCTAATTCTCACTATGAtGAATGTTCCTGTTAC-3’, 5’-CATTCaTCATAGTGAGAATTAGGTGC-3’ that change the codon from GAG to GAT (The changed nucleotides for the overlapping primers are marked as lowercase letters). Primers were made by Evrogen (Moscow, Russia). The changed PCR product was then cloned into the same backbone.
Tet-on system was set up to make transcription of the gene of interest to be inducible (Gossen & Bujard, 1992; Gossen et al., 1995). rtTA (tetracycline transactivator), CMV promoter, TRE (Tet Response Element) promoter P2A and RFP were amplified from different plasmids. Influenza A neuraminidase and sialidase from S. typhimurium were inserted under control of TRE-promoter. Neuraminidase was amplified from the plasmid pNAHA (Plasmid # 44169, Addgene) and sialidase S. typhimurium was amplified from pHAGE-CMV-Sal.t.Sia-RFP. All amplified products were cloned in the pHAGE backbone in the following order: TRE-infNA/Sal.t.Sia-P2A-RFP-CMV-rtta by restriction cloning (Supplementary Figure 1B). The plasmid pTagBFP encoding BFP (blue fluorescent protein) was used for cotransfection to approximately estimate effectiveness of transfection before addition of the doxycycline.
Cells were treated with Doxycycline (D9891, Sigma; St Louis, MI, USA) at 24 hours after transfection at a concentration of 0.5µg/ml. Expression of the RFP reporter and activity of genes-enzymes were analyzed at 48 hours after Doxycycline addition.
MDCK and HEK293 cell lines were cultured according to ATCC recommendations. Cells were maintained at 37°C with 5% CO2. Cells were transfected with TurboFect reagent (R0531, Thermo Fisher; Waltham, MA, USA) according to the manufacturer recommendations. The fluorescence microscope Axio observer (Zeiss, Oberkochen, Germany) with the standard set of filters was used for visualization of fluorescence. S3e BioRad sorter (Hercules, CA, USA) was used for fluorescence activated cell sorting and flow cytometry. Standard flow cytometry analysis was carried out using FlowJo v10.4 software.
FITC fluorescence in Lectin binding assay was measured by FACS and quantified as an average mean fluorescence intensity (MFI). Values show the means ± SD of triplicate results from representative experiments. Three independent experiments were carried out for each experimental case. Student’s t-test was used to determine the level of statistical significance. Statistical analysis was perform in Microsoft Excel 2016
Lentivirus packaging was performed according to the described protocol [see Protocol 1: Lentivirus Packaging by 293T Transfection from CReM].
In order to determine a viral titre, different aliquots of supernatant were added to HEK293 cells in the presence of 4 μg/ml polybrene (Hexadimethrine bromide, H9268, Sigma). Infection of six 10-fold serial dilutions was performed in 96-well plates. The medium was replaced 12 hours after infection. Three serial infections were performed. At 48 hours after infection, transduction effectiveness was calculated by FACS analysis. The viral titer corresponded to the number of colonies developed at the highest dilution.
The supernatant of the known titre was used for viral transduction of the MDCK cell line. On the first day about 1.0 x 10-4 MDCK cells were seeded into 6-well plates. The cells were incubated 18–20 hours at 37°C in a humidified incubator in an atmosphere of 5–7% CO2. Upon transduction the confluency around 30–50% was estimated. Next, the appropriate volumes of unconcentrated virus and polybrene with concentration 5ug/ml were added to each well. After 48 hours, cells were analyzed using Flow cytometry to define the percentage of infected/fluorescent cells. Lentiviral transduction efficiency was calculated as (the number of RFP-positive cells/the total number of cells counted × 100) and the number of transduced cells (transduction efficiency (%) × the total number of RFP recovered/100).
Genomic DNA was isolated using Wizard® Genomic DNA Purification Kit (A1120, Promega, Madison, WI, USA) according the manufacturer protocol.
Lectin binding assay was used for the cell surface SA detection. For lectin binding assay we used Maackia amurensis (MA) lectins that are specific for α (2–3)-bound SA. The lectin were conjugated with the FITC fluorophore (21761036-1 (510183), bio-WORLD; Dublin, OH, USA). The assay was made for HEK293 and MDCK cells. The experiment was setup in 48-well plates. The cells expressing a desired construct were washed three times with PBS. Then cells were incubated with MA lectins (1:100) about 1 hour and three times washed with PBS. Hoechst (RRID:AB_2651133, Thermo Fisher) was added for visualization of the nuclei for 5 min at recommended concentration with the subsequent washing with PBS.
Sialidases vary in enzyme kinetic and substrate specificity to the type of the SA linkage. Several sialidases from bacteria, Influenza virus and vertebrate were selected for our study. We were mostly interested in α(2–3) specialized sialidases, because avian Influenza prefers this type of linkage (Ito, 2000). However the broad substrate specificity was also an area of interest. Regarding enzyme kinetics, sialidases with various levels of activity were useful for us in order to combine them with tissue-specific promoters and find an optimal level of desialilation. The optimal sialic acids level on the cells surface prevents penetration of the virus into the cell and does not affect cell function.
Substrate specificity of bacterial sialidases is very diverse, in terms of type of sialic acid linkage and enzyme kinetics. We selected two different bacterial sialidases: Salmonella typhimurium sialidase, which specializes in cleavage of α(2–3) sialic acid residues (Hoyer et al., 1991; Rogerieux et al., 1993) and Actinomyces viscosus sialidase, which has a substrate specificity to both α (2–3) and α (2–6), but preferentially cleaves the α (2–6) linkage (Teufel et al., 1989). We used sequence of the catalytic domains of the sialidases and added to their N-terminal sequences the transmembrane domain with the stem loop of Influenza NA transmembrane protein in order to target the proteins to the membrane and demonstrate its catalytic activity outside of the cell.
The human Influenza neuraminidase (sialidase) has been taken for analysis as a broad-spectrum sialidase. Unlike human HA that preferentially recognises α (2–6), the viral NA of H1N1 has cleavage activity to both (2-6) and α(2–3) types of linkages.
Exogenous overexpression of a vertebrate membrane sialidase also would be interesting to analyze. The sialidases are poorly investigated, but some experimental data exists for human membrane neuraminidase hNeu3 (Monti et al., 2000; Zhang et al., 2010) and the enzyme was selected for cloning.
All cloned genetic constructs were firstly tested by transient expression in the HEK293 cell line that has glycosylation patterns including α (2–3) and α (2–6) sialic acid linkages (Picanco-Castro et al., 2013). All plasmids had the RFP reporter to mark the cells that express a sialidase. Using lectin binding assays it has been shown that the cells expressing the sialidase from S. typhimurium have decreased level of SA. Expression of the sialidase from A. viscosus did not affect the α (2–3) sialylation level, probably because it is more specific to the α (2,6) sialic acid linkages. However, it has previously been shown that this sialidase effectively removes both α (2,3)- and α (2,6)-linked SA from the cell surface (Malakhov et al., 2006). Human neuraminidase hNEU3 also has not shown activity towards α (2–3)-linked SA. We suppose that it might be related to unsuitable pH conditions. Cultivation in DMEM in 5% CO2 provides pH in range of 7,6–7,7. Earlier, hNeu3 activity was demonstrated in the pH range of 2.8–6.6 (Monti et al., 2000). In the case of influenza A neuraminidase, sialylation level decreased, not only in RFP-positive cells, but also in RFP-negative cells, which are not supposed to express the enzyme (Figure 2A). According to the flow cytometry analysis, the most significant α (2–3)-linked SA cleavage was in the case of Influenza A neuraminidase expression (Figure 2B). Representative histograms from three independent repeats are shown. Lectin binding assay was quantified as fluorescence and calculated as a mean fluorescence intensity - MFI (±SD): d_infNA-RFP - 103.437 (±3.568), Act.v.Sia-RFP - 181.401(±7.988), Sal.t.Sia-RFP - 7.341(±1.323), infNA-RFP - 4.725(±983), hNeu3 - 27.379(±2.231).
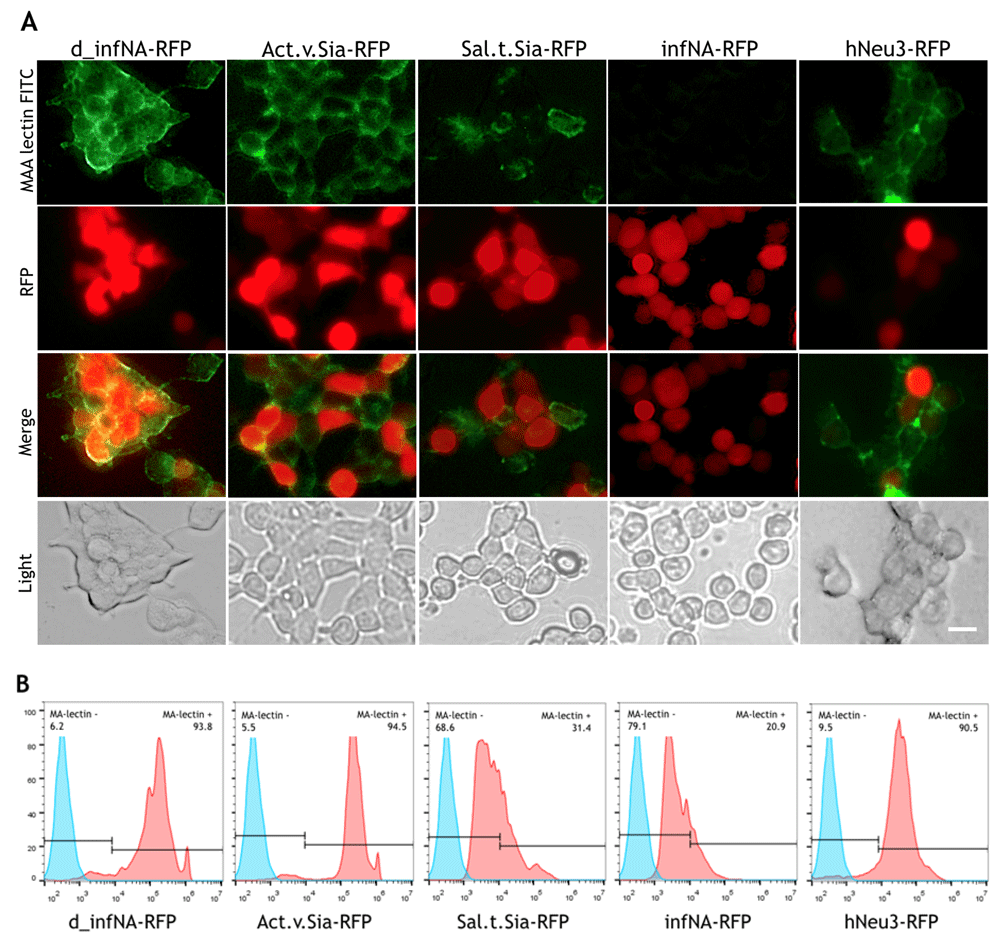
Genetic constructs were transiently expressed in the HEK293 cell line, and sialidase activity was evaluated using a lectin binding assay with FITC-labelled Maackia amurensis lectins. The plasmid with catalytically inactive neuraminidase was used as the negative control. (A) Fluorescent microscopy analysis. Scale bar = 10 um. (B) FACS analysis.
A responsive transcription of a sialidase gene gives an opportunity to induce the gene expression in response to a molecule addition. In our case we used Tetracycline-Controlled Transcriptional Activation where transcription is reversibly turned on in the presence of the antibiotic tetracycline derivative doxycycline. The method can be a useful model for studying viral infection in cell culture and especially for use in genetically modified organisms because constitutive exogenous gene expression could have a potential harm.
For tetracycline-inducible expression we selected only sialidases that demonstrated significant α (2–3) catalytic activity in the previous experiment with transient expression under CMV promoter (infNA-RFP and Sal.t.Sia-RFP). The plasmid coding BFP under constitutive CMV promoter was used for estimation of transfection effectiveness. It was shown that upon treatment of cells with doxycycline the sialidases expression switched on and as a result the α (2–3) sialylation level decreased (Figure 3A, B). From flow cytometry analysis representative histograms from three repeats are shown. MFI (±SD) was calculated: infNA-RFP, +dox - 1.468 (±439), Sal.t.Sia-RFP, +dox - 6.437(±1.043), d_infNA-RFP, +dox - 83.849(±2.983).
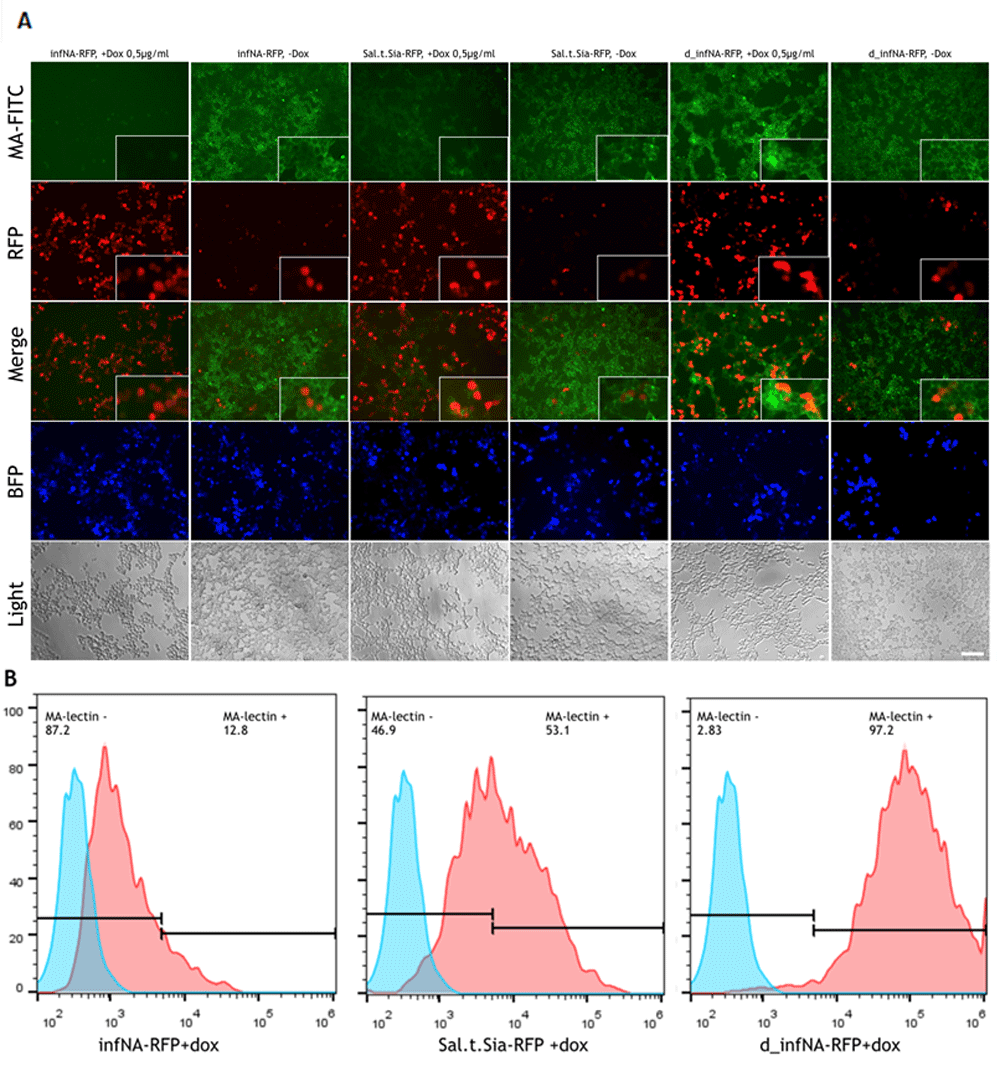
Gene expression was evaluated in the HEK293 cell line after doxycycline induction, using a lectin binding assay with FITC-labelled Maackia amurensis lectins. The plasmid with catalytically inactive neuraminidase was used as the negative control. (A) The plasmid coding BFP was used to estimate the effectiveness of transfection. Fluorescent microscopy analysis. Scale bar = 50 um. (B) FACS analysis.
The MDCK cell line is used for propagation of influenza viruses. To obtain cell lines that stably express exogenous genes coding for different sialidases we transduced MDCK cells with lentiviruses. The expressing construct was integrated into the genome by lentiviral transduction. The integration was confirmed with PCR (Supplementary Figures 2 and 3) using primers from Table 1. The effect of expression of sialidases was studied by testing the reactivity of cells with sialic acid linkage-specific lectins.
The results confirmed the previously obtained data with the HEK293 cell line. The cells expressing S. typhimyrium sialidase and Influenza A neuraminidase had decreased level of the surface α (2–3) SA. However, the decrease in the SA level in the MDCK was less significant compared with the HEK293 cell line (Figure 4B). Representative histograms from three technical repeats are shown. MFI was calculated: d_infNA-RFP - 38.120(±2.322), Act.v.Sia-RFP - 22.750(±3.175), Sal.t.Sia-RFP - 21.732(±4.598), infNA-RFP - 7.290(±1.934), RFP-NA - 12.798(±3.453). Similarly with the experiments on the HEK293 cell line, in the case of infNA-RFP expression α (2–3) SA were absent not only in the RFP-positive cells but in the RFP-negative cells as well. A possible explanation of the evidence could be a ‘slipstream’ translocation in a polycistronic vector of the P2A downstream protein without a signal of localization that is RFP in our case (de Felipe et al., 2010). An alternatively explanation being when a ribosome encounters 2A within an open reading frame the synthesis of a specific peptide bond could be “skipped”. This results in termination of translation at the end of 2A peptide (Donnelly et al., 2001). It means that RFP is not expressed and can explain the absence of sialic acids in some RFP-negative cells. The effects were not observed in the case of other sialidases. Usage of a longer sequence for P2A (with a favorable upstream sequence composition) or modifying the order of proteins could solve the problem. When we changed the order of the RFP and the infNA sequences (RFP-P2A-infNA), the result of lectin binding assay was as expected: RFP-positive cells showed decreased level of α (2–3) SA (Figure 4A). Nevertheless, in the case of inverted position of the neuraminidase the enzyme activity was less pronounced. It can be explained by the fact that a protein located at the second position can be slightly less expressed in a polycistronic construct with a 2A peptide (Liu et al., 2017). In the case of the S. typhimyrium sialidase expression (Sal.t.Sia-RFP) the ‘slipstream’ effect was not observed.
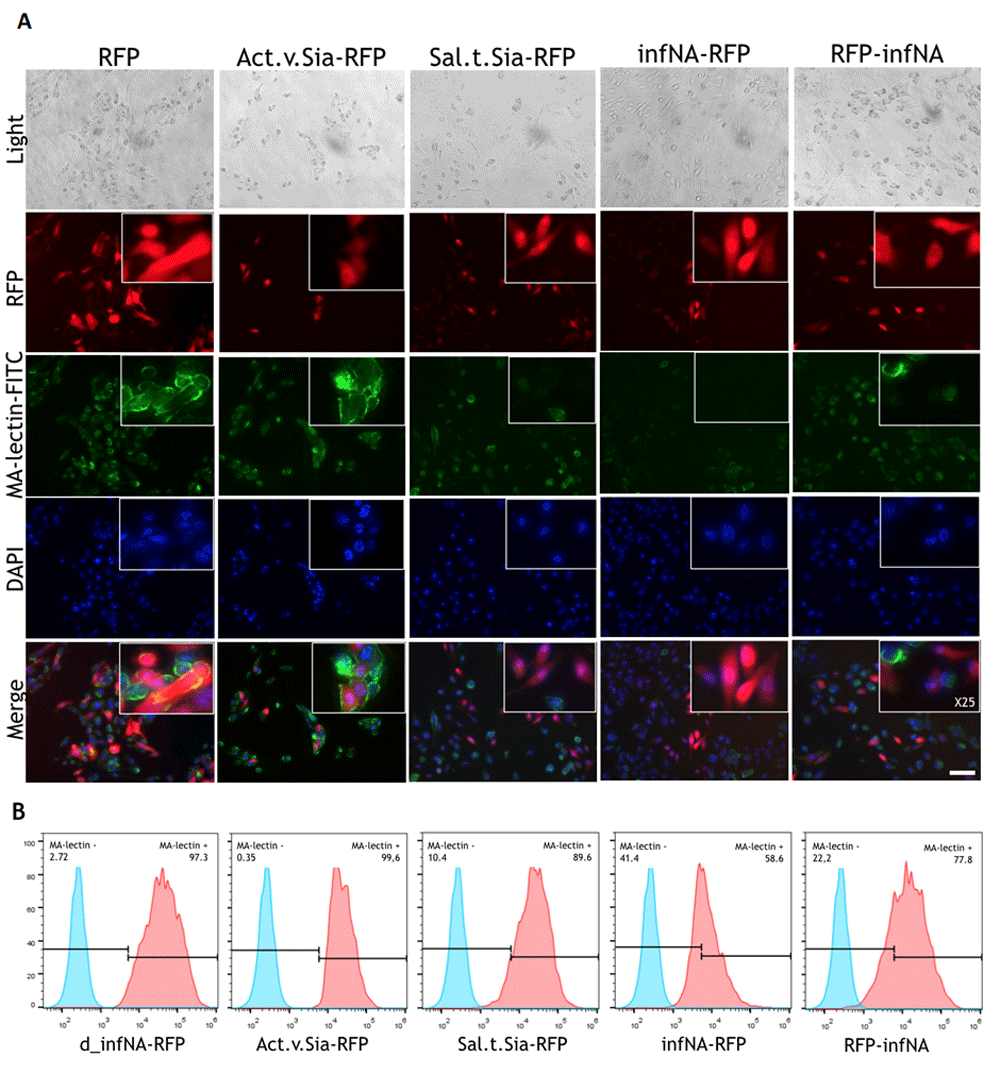
MDCK cells were transduced with lentiviruses. Lectin binding assay was made with FITC-labelled Maackia amurensis lectins. The plasmid with catalytically inactive neuraminidase was used as the negative control. (A) Fluorescent microscopy analysis. Scale bar = 50 um. (B) FACS analysis.
Sialic acid is the sole receptor of Influenza A virus (Matrosovich et al., 2013). Therefore the removal of sialic acids from the cell surface might be a powerful defence strategy against Influenza virus. In the current study we take a variety of sialidases from different sources and compared their activity. We demonstrated that expression of the sialidase catalytic domain from Salmonella typhimurium fused with the transmembrane domain and human Influenza A neuraminidase effectively removes α (2–3)-sialic residues on the cell surface under physiological conditions.
In our study, about 70–80% of the surface SA were removed by the membrane sialidases. It has been reported that the virus binding could still occur with such sialylation level, as has been shown in the MDCK cell line, but amplification of Influenza virus was still inhibited (Stray et al., 2000), therefore complete elimination of SA is perhaps unnecessary. Neuraminidase had higher cleavage activity against α (2–3)-linked sialic acids than sialidase from S. typhimyrium according to our results. An optimal level of desialilation can be established in the tissue when combining genetic constructs with varying sialidase activity and tissue-specific promoters.
Stable expression of sialidases potentially could disrupt physiological functions required for proper glycosylation (Gutierrez et al., 1987; Michalek et al., 1988; Pangburn et al., 2000; Varki, 1992). Inducible expression may be more suitable in this case. The Tet-on inducible system allowed robust expression and functioning of genetic constructs encoding sialidases after the inductor addition. The level of sialilation will return to the previous level during a period of time after withdrawal of doxycycline. The required time depends on the sialic acid turnover rate.
It has been shown that sialidase treatment does not affect the properties of respiratory mucus, nor did it affect the normal mucus transport activity on ciliated epithelium (King et al., 1974; Meyer et al., 1975). Thus, temporary desialilation of the epithelial surface should not cause problems.
Our current preliminary in vitro data indicates that sialidases from Salmonella typhimurium and neuraminidase from Influenza A virus could be the potential candidates that provide antiviral defense against avian Influenza virus. Since sialidases target cellular receptors but not a viral gene product, the chance of influenza viruses developing resistance is low.
Data underlying this study is available from Dataverse - doi:10.7910/DVN/UCDPX3 (Antonova, 2018).
This work has completed with the financial support of Russian Science Foundation within project No. 16-16-04094.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
No
Are the conclusions drawn adequately supported by the results?
No
References
1. Nicholls JM, Moss RB, Haslam SM: The use of sialidase therapy for respiratory viral infections.Antiviral Res. 2013; 98 (3): 401-9 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Influenza molecular virology. Transgenic livestock.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Influenza
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 16 May 18 |
read | read |
|
Version 1 19 Feb 18 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)