Keywords
Cytoscape, CyTargetLinker, network extension, network visualization, regulatory networks, data integration
Cytoscape, CyTargetLinker, network extension, network visualization, regulatory networks, data integration
We updated the website (content and URLs) and tutorials to make sure all information is up-to-date.
We added some additional information regarding the API command used in the use cases.
See the authors' detailed response to the review by John H. Morris
See the authors' detailed response to the review by Alex Gutteridge
The CyTargetLinker app provides a flexible and simple way to extend networks in Cytoscape1 with links to (prior) knowledge from external sources. Since its first release in 20132, CyTargetLinker has been downloaded more than 19,000 times and used in numerous studies. These applications in biological studies range from the creation of a microRNA-gene association network for lipid diseases3 or Alzheimer’s disease4 to the application of algorithms for drug sensitivity prediction5.
While the app was originally intended to be used for the extension of biological networks with regulatory interactions, researchers have used CyTargetLinker to integrate knowledge about many different types of relationships (e.g. pathway associations and disease annotations). Therefore, we renamed the previously used Regulatory Interaction Networks (RegINs) to linksets to make the broader applicability more explicit. Moreover, the generation of linksets, either manually or in an automated manner, has become more user-friendly.
In this new version of CyTargetLinker, we introduce an automation feature that allows programmatic access to the app functionality. In the Results section, we present three use cases that highlight the app’s purpose, how it can be easily combined with other Cytoscape apps and the advantages of the automation. Whereas the first two use cases have a biological nature, the third use case demonstrates the broader applicability with a non-biological example. Additionally, the website and tutorials have been updated and restructured (https://cytargetlinker.github.io/).
The newest version of CyTargetLinker (4.0.0+) was developed for Cytoscape (3.6.0+) which introduces a new interface for automation that can make apps callable as services by the Cytoscape Command scripts, Python and R. This promotes open and reproducible data analysis, and simple integration with other apps. CyTargetLinker can be installed through the Cytoscape app store.
On the CyTargetLinker website, we provide a variety of linksets for regulatory interactions, pathway associations and disease annotations (https://cytargetlinker.github.io/pages/linksets). Additionally, we deliver a simple Java program to convert tab delimited text files into XGMML linksets that can be used with CyTargetLinker (https://github.com/CyTargetLinker/linksetCreator). Using BridgeDb6, a framework for finding and mapping database identifiers, the script enables the support of multiple identifier systems for biological entities.
While CyTargetLinker can still be used through the Cytoscape graphical user interface (see online tutorials), we would like to highlight the novel application programming interface (API) that allows the programmatic execution of the app’s functionality.
CyTargetLinker provides a set of API methods to automise the extension of networks (Table 1). The key function is the “extend” function, which parses the provided linksets and extracts relevant interactions for the selected network. The user can then choose to use the CyTargetLinker visual style and the force-directed layout. Often, users want to integrate knowledge for the same interaction type from different resources. With the “filterOverlap” function, users can visualise only those interactions that are supported by multiple resources.
List of the API methods of CyTargetLinker, their parameters and a general description.
The broad applicability of CyTargetLinker will be demonstrated in three different use cases. The focus lies on the automation of the analysis and the R scripts for each use case are provided in the automation repository on GitHub. We chose to present two biological and one non-biological use cases to demonstrate the flexibility of the app.
Use case 1: Investigating drug-targets and disease associations for a Rett syndrome protein-protein interaction network. Rett syndrome is a rare disease caused by a mutation in the methyl-CpG-binding protein 2 (MECP2) gene7. In this use case, we used the stringApp8 of Cytoscape to create a protein-protein interaction (PPI) network for Rett syndrome (Disease Query). The PPI network is then extended using CyTargetLinker with compound-target interactions from ChEMBL9,10 and disease-gene associations from a manually curated subset for rare diseases from OMIM11. ChEMBL is an open online bioactivity database containing information about compounds, their bioactivity and their possible targets (including proteins). OMIM is a comprehensive collection of human genetic phenotypes and their associated human genes. First, the stringApp was used to create a Rett syndrome PPI (query=“Rett syndrome”, cutoff=0.4, limit=20). Using CyTargetLinker, the network was extended with 37 compound-target interactions from ChEMBL and 18 gene-disease associations from OMIM (see Figure 1).
The following API command was used to extend the network with compound-target and disease-gene information:
cytargetlinker extend idAttribute=”display name” linkSetFiles=”../LinkSets/chembl_23_hsa_20180126.xgmml, ../LinkSets/omim-rare-disease-has-20180411.xgmml” network=current
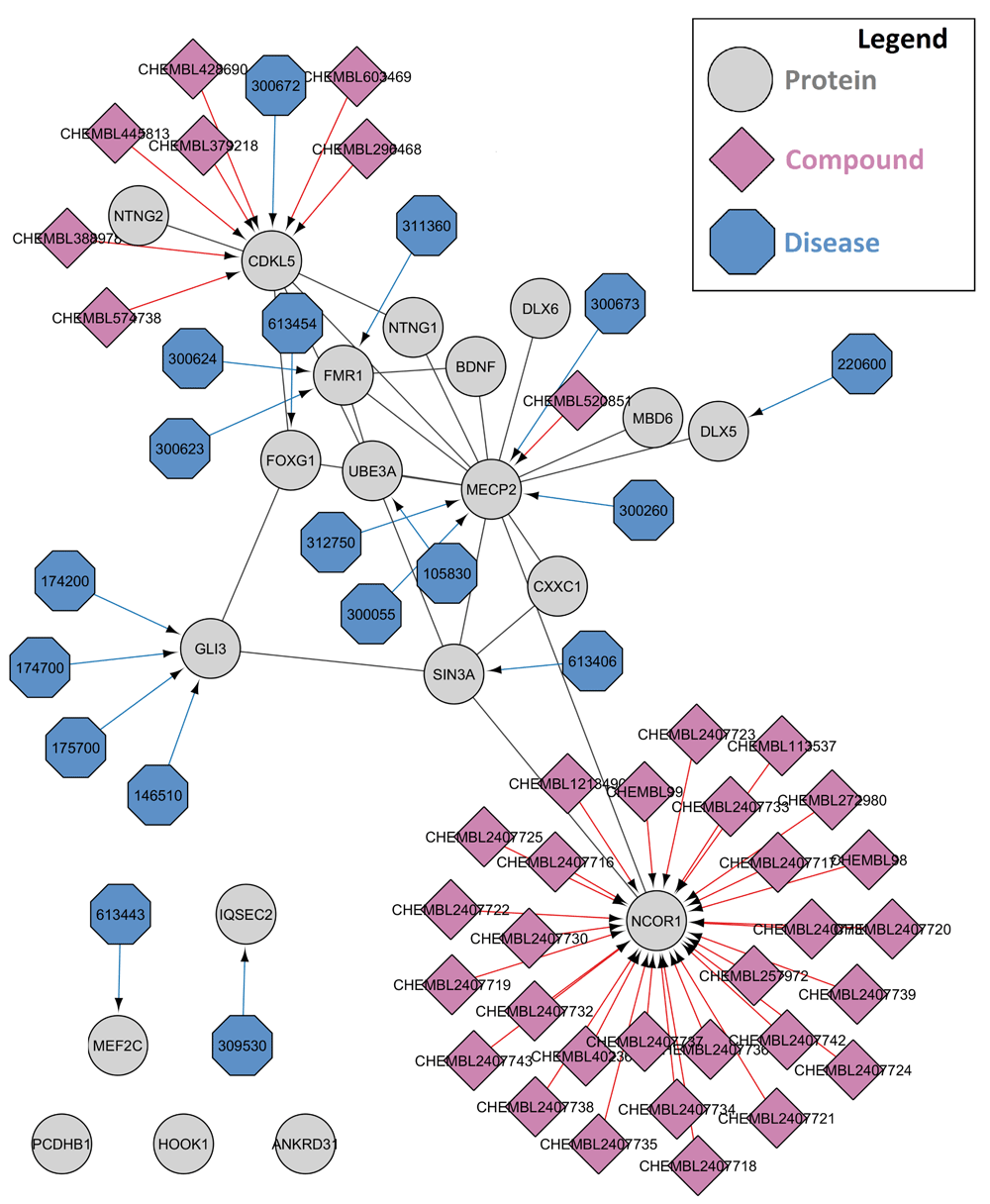
The protein-protein interaction network for Rett syndrome was created using the disease query option of the stringApp for Cytoscape. The proteins are represented as gray circles. Then, CyTargetLinker was used to extend the network with compounds from ChEMBL (purple diamonds) and disease annotations from OMIM (blue octagons).
Use case 2: Pathway associations for differentially expressed genes in Rett syndrome. For this use case, we selected a list of differentially expressed genes in the Purkinje cells located in the cerebellar cortex of the brain of a Mecp2−/y mouse model12,13 for Rett syndrome. Next, we investigated in which biological processes these altered genes are involved. Using the pathway annotations from the WikiPathways database14, CyTargetLinker adds the pathway information and creates a pathway-gene network.
From the dataset, we extracted 65 genes with an absolute log2 fold change larger than 1. Only 16 genes are present in one or more pathways of the curated mouse pathway collection from WikiPathways. Figure 2 shows the resulting gene-pathway network. Genes without pathway annotations have been removed. Differential gene expression is shown on the gene nodes (blue = down, red = up) and green border color of the pathway nodes indicates that the pathway has been identified as significantly affected through overrepresentation analysis in the pathway analysis tool PathVisio15.
The following API command was used to extend the network with pathway information:
cytargetlinker extend idAttribute=”shared name” linkSetFiles=”../LinkSets/wikipathways-mm-20180410.xgmml” network=current direction=SOURCES
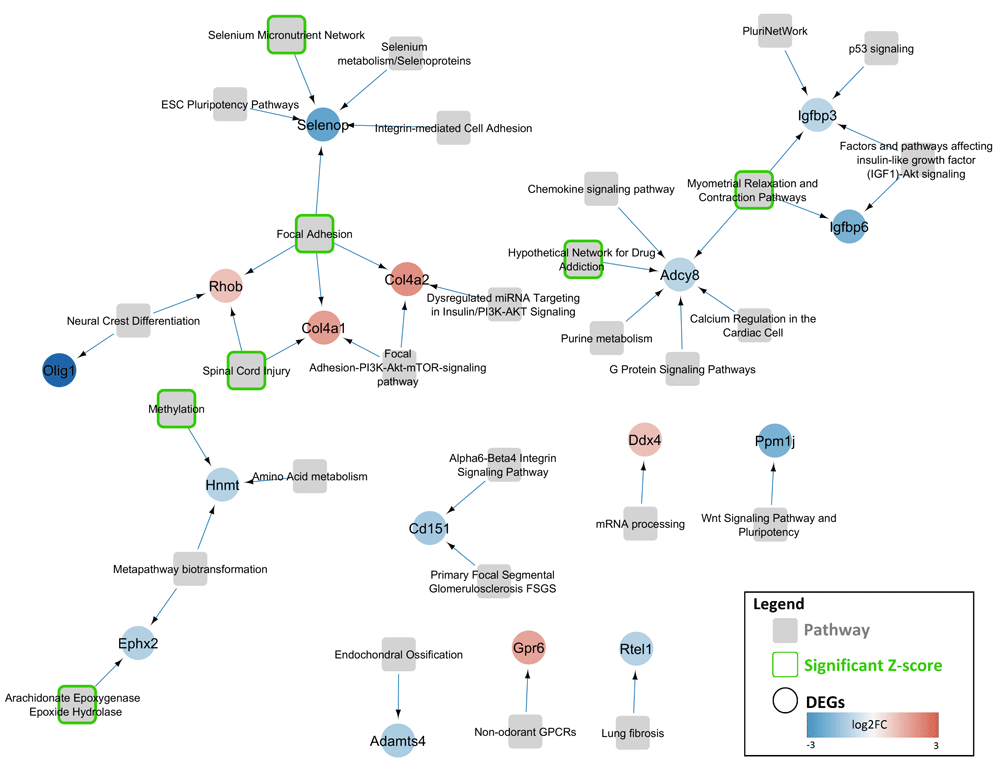
DEGs in a mouse model for Rett syndrome (Mecp2−/y mice) were selected and imported in Cytoscape (circular nodes). The genes are colored based on changes in gene expression (blue=down-regulated, red=up-regulated). Thereafter, gene-pathway associations from WikiPathways were added and the pathways are shown as gray rectangles. Genes without pathway annotations have been removed. The green border color indicates pathways that are significantly altered based on over-representation analysis in PathVisio (Z-Score > 1.96).
Use case 3: Author-publication-journal network. This example uses two custom made linksets for author-article and article-journal relationships from Wikidata16–18. After loading the initial five author nodes in Cytoscape, we performed a two-step extension with CyTargetLinker. We first added publications from the author-article linkset and then the journals from the article-journal linkset, see Figure 3. Author nodes are colored in gray, articles in yellow and journals in green. The network clearly shows the collaborations and diversity between the authors. Layout and visual style was slightly adapted manually in the graphical user interface to improve the readability of the network.
The following API commands were used to extend the network first with publication and then with journal information:
cytargetlinker extend idAttribute=”shared name” linkSetFiles=”../LinkSets/publications.xgmml” network=current cytargetlinker extend idAttribute=”shared name” linkSetFiles=”../LinkSets/journals.xgmml” network=current
One of the major challenges in science is the reproducibility of results presented in articles. Besides the challenges in reproducibility of experiments, the computational analyses are also often unclear and insufficiently described19. Automation of analysis workflows enables researchers to share the details of their computational analyses and enables simple reproducibility of the results.
Here, we introduce the new version of the CyTargetLinker app, which provides full programmatic execution of the functions from within Cytoscape (command line tool) and external tools (e.g. R, Jupyter, Python, etc). The network extension is therefore reproducible and repeatable with other input data. Consequently, users can build scripts that run common analysis workflows and combine CyTargetLinker with other apps, as shown in Use case 1 (stringApp). The integration of CyTargetLinker in Cytoscape gives access to a powerful set of visualization options, as demonstrated in use case 2.
As part of the Cytoscape tutorials collection for online presentations, we developed a CyTargetLinker tutorial presentation using Reveal.js. This tutorial presentation can be reused and adapted for specific teaching activities. Together with our tutorials for the Cytoscape desktop application and the automation example scripts, relevant documentation for users is provided to get familiar with the functionality of CyTargetLinker.
The generic nature of CyTargetLinker has been highlighted by renaming RegINs to linksets, and we now provide a variety of different linksets on our website. The XGMML structure of the linksets is simple, instructions how to create them from tab-delimited text files are available, and CyTargetLinker could therefore be used for non-biological networks as well (shown in use case 3).
In this paper, we highlight the latest update of the CyTargetLinker app for Cytoscape and its new automation feature. The ability to programmatically execute the app’s functionality opens up the possibility to build complex workflows that are repeatable and reproducible. We also explored the broader applicability of the app besides the originally intended use for regulatory network extension. Due to the flexible design of the app and the linksets, we are now also showcasing other use cases, including non-biological networks.
Linksets, tutorials and link to source code (app and linkset creator) are available from the CyTargetlinker app website: https://cytargetlinker.github.io/.
1. The app is available from the Cytoscape app store: http://apps.cytoscape.org/apps/cytargetlinker.
2. Link to source code: https://github.com/CyTargetLinker/cytargetlinker.
3. Archived source code at time of publication: https://doi.org/10.5281/zenodo.336238920.
4. Tutorials including R code for use cases: https://github.com/CyTargetLinker/cytargetlinker-automation.
5. Software license: https://www.apache.org/licenses/LICENSE-2.0.
This project has been co-financed by the Dutch Province of Limburg and ELIXIR, the European research infrastructure for life-science data.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We would like to thank the Cytoscape developer team, especially Barry Demchak, Scooter Morris and Alex Pico, for the support during the automation feature implementation.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Network Biology
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Network Biology
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 13 Aug 19 |
read | |
|
Version 1 14 Jun 18 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)