Keywords
Consensus Diversity Plot, compound databases, data mining, diversity, natural products, functional groups, in silico
This article is included in the Cheminformatics gateway.
This article is included in the Mathematical, Physical, and Computational Sciences collection.
Consensus Diversity Plot, compound databases, data mining, diversity, natural products, functional groups, in silico
Natural product-based drug discovery continues to be an important part of drug discovery. Recently, the synergy between natural product research with molecular modeling and chemoinformatics is gaining importance, speeding up the drug discovery process1,2. As part of these synergistic efforts, curated databases of natural products have an important role as they are major tools for data mining, hypothesis generation, and starting points of virtual screening. There are several databases of natural products in the public domain as reviewed recently3. Our research group has reported initial efforts to assemble a database of natural products from Mexico called BIOFACQUIM4. As part of that work, it was reported an initial scaffold content, and chemical space diversity, and coverage. However, detailed functional group (FG) content analysis, which has been proven to be valuable to characterize compound databases5, in particular from natural sources6, has not been reported for BIOFACQUIM. One of the main reasons is that most of the currently available software employed for identification of functional groups rely on a predefined set of substructures, even when it has been established that one of the major features that discriminate natural products from synthetic compounds are their unique functional groups.
Herein, we report a functional group content analysis of an updated version of BIOFACQUIM. We employed a validated and novel algorithm that identifies all functional groups in a molecule. As part of the analysis and to compare the results of BIOFACQUIM we also discuss the functional group contents of other large and related databases in the public domain, namely ChEMBL 257 and a herein assembled collection of natural products (NPs) with 169,839 compounds.
As described elsewhere, the first version of BIOFACQUIM was developed as a proof-of-concept database applying several filters to include compounds4. Briefly, the database was focused on natural products published between 2000 and 2018 by research groups in a major Mexican institution in eight indexed journals. As additional criteria for inclusion of compounds and to increase the quality and reliability of the contents of the database, the articles should have described the procedure for the isolation, purification, and characterization of the natural product. In this work, we decided to expand the contents of the BIOFACQUIM database to further explore the diversity of natural products from Mexico.
The second version of BIOFACQUIM was assembled using the same methodology described to develop the first version4. To achieve the objective of being representative of Mexico, it was decided to augment the number of Mexican institutions (universities, research laboratories, and research centers) publishing information of novel natural products. For the new version of the database, the same procedure for the curation was performed4, using Molecular Operating Environment (MOE) software. A similar procedure can be done using RDKit. In addition, it was ensured to store in the database, natural products that were collected in Mexico, only. The updated and curated version of BIOFACQUIM contains 531 compounds.
Table 1 summarizes the information of BIOFACQUIM and other major compound databases used in this work as reference: ChEMBL 25 as a representative example of the biologically tested chemical space with 1,667,893 unique compounds; and a collection of known natural products with a total of 169,839 molecules. The reference natural product collection was assembled from three general and publicly available natural products databases: the Universal Natural Products Database (UNPD)8, the Natural Products Atlas9 and Natural Products in PubChem Substance Database10. The data sets were curated using the same procedure. Briefly, compounds consisting of multiple components (e.g. salts) were split and the largest component (defined by the number of heavy atoms) was retained, as long as the number of heavy atoms of the second largest component was less than 70% of the largest component. If this was not the case, the compounds were removed from the dataset, unless the two largest components were identical. Single component compounds as well as the retained component of multiple component compounds were neutralized and canonical simplified molecular-input line-entry system (SMILES) (ignoring stereochemistry information) were generated as molecular representation. Compounds consisting of any element other than H, B, C, N, O, F, Si, P, S, Cl, Se, Br and I, as well as compounds with valence errors, were removed from the dataset. Duplicate structures in the context of each database were also removed. The entire process was performed by using the open source cheminformatics toolkit RDKit.
Overlap of BIOFACQUIM with the databases selected as reference was assessed in terms of three different structural levels: compounds, scaffolds, and functional groups. Compound overlap was determined in terms of canonical SMILES. For scaffold comparison we use the definition proposed by Bemis and Murcko11 as implemented in RDKit, while for functional group overlap we selected the recently published definition and implementation suggested by Ertl5. For each structural level, we identified the unique structures belonging to each dataset as well as those belonging to two or three of them.
For the functional group content analysis we selected the algorithm recently described by Ertl5, which is able to identify all functional groups in a molecule based on an iterative marching through its atoms. In short, the proposed algorithm identifies all heteroatoms in a molecule, all atoms connected by multiple bonds as well as the atoms in oxirane, aziridine, and thiirane rings. Afterwards, all connected atoms are joined together to form a functional group. Single aromatic heteroatoms are retained only if they are connected to an additional aliphatic functionality. Finally, a generalization scheme is applied in which for a defined list of common FGs, information about the parent carbon is retained (e.g. to differentiate between alcohols and phenols) as well as hydrogen atoms (e.g. to differentiate between aldehydes and ketones). The method is fully described in 5. An open source version of this algorithm is available for Python (https://github.com/rdkit/rdkit/tree/master/Contrib/IFG); however, it does not cover the generalization scheme proposed originally. To this end and based on the code available, we implemented with RDKit a fragmentation approach considering the retainment of parent carbons and hydrogens proposed originally, were the remaining carbon atoms are replaced by dummy atoms. This implementation works over a SMILES string and returns a list with the canonical SMILES of the FGs identified in the molecule. The code is freely available at GitHub (https://github.com/DIFACQUIM/IFG_General). After determining the FGs content of the different datasets, we compare the proportion of the most frequent FGs at each library.
In order to generate a visual representation of the chemical space covered by the analyzed databases, six molecular properties of pharmaceutical interest were computed for each unique compound: averaged molecular weight (AMW) partition coefficient octanol/water (SlogP), number of hydrogen bond donors (HBD), number of hydrogen bond acceptors (HBA), number of rotatable bonds (RB), and topological polar surface area (TPSA). The correlation matrix was computed and a visual representation of the chemical space was generated by using the two properties with the lowest correlation: AMW and SlogP. All calculations were done with RDKit.
The “global” or total diversity of the datasets was analyzed through the Consensus Diversity (CD) Plot12 that was designed to capture, in a two-dimensional graph, the diversity of chemical libraries considering different and complementary criteria including molecular fingerprints, molecular scaffolds, physicochemical properties, and size. For this work, scaffold diversity was assessed as the area under the cyclic system retrieval curve and the fraction of chemotypes that covers 50% of the dataset (F50). The median of the lower triangle from the pairwise similarity matrix computed as the Tanimoto coefficient of both MACCS keys (166-bits) fingerprint and Morgan fingerprint with radius 2 (Morgan2), were used as molecular fingerprint-based diversity. The mean distance of the lower triangle of the pairwise distance matrix computed as the Euclidean distance of the six molecular properties scaled (mean 0 and unit variance) and the number of compounds in each dataset were selected as measures of the physicochemical properties- and size- based diversity, respectively. The CD Plot was constructed using R.
As described in the Methods section, the updated BIOFACQUIM database contains the chemical structure of 531 compounds, all collected from Mexico. As with the first version4, each molecule is annotated with information of the chemical structure, the original source of the information (Digital Object Identifier, DOI, to reference paper), kingdom, genus, and species of the plant the natural product was isolated, place of recollection (city and state), and activity value of the reported biological activity. From the original dataset containing 423 compounds, 40 were discarded since they were not collected in Mexico, which means an increase of 148 unique compounds compared to the previous release of the database.
To assess the chemical space not covered by ChEMBL and NPs but by BIOFACQUIM, we characterized the structural content of the three datasets in terms of unique compounds, scaffolds and functional groups and determined the overlap among them. Figure 1 depicts Venn diagrams showing the overlap among those datasets. It should be noted that despite its small size in comparison with ChEMBL and NPs, 16.1% of compounds present in BIOFACQUIM have not been reported in those other major datasets, as well as 11.3% of its scaffolds and 6.3% of its FGs. Another remarkable observation is the fact that most of the overlap of BIOFACQUIM with the other datasets involves NPs, either alone or in combination with ChEMBL, being 79.7%, 85.5%, and 93.8% for compounds, scaffolds and FGs, respectively.
A systematic analysis of functional groups was carried out over BIOFACQUIM and the two datasets selected as reference. 80, 4300, and 12,677 functional groups were identified in BIOFACQUIM, NPs and ChEMBL datasets, respectively (the overlap between them is shown in Figure 1). From the total number of functional groups present in each dataset, only 12, 14, and 23 were present in at least 1% of the corresponding library (15.0%, 0.3% and 0.2%, respectively) while 47, 2207, and 5853 (58.7%, 51.3% and 46.17%, respectively) were singletons. This result is consistent with the typical power law observed in other databases6. The most frequent FGs present in BIOFACQUIM are oxygen-containing FGs, being the phenolic hydroxyl group (41.4%), followed by the alcohol hydroxyl group (40.0%), ether (39.8%), alkene (26.6%) and ester (25.8%), which although in a different order, are the most frequent FGs in the herein assembled NPs collection and in other natural product libraries6. This is in contrast to ChEMBL in which only ether is part of the most frequent FGs while the rest of them are nitrogen containing FGs. The complete results of the FGs found of the datasets is included as Extended data (Supplementary File 1). Remarkably, even with the relatively small size of BIOFACQUIM, five compounds with unique FGs were identified, three of them isolated from Mexican plants, two from Salvia ballotiflora (FQNP204 and FQNP211)13 and one from Heterotheca inuloides (FQNP480)14, while the other two correspond to fungal metabolites from Malbranchea aurantiaca (FQNP435)15 isolated from bat guano, and from the endophytic fungus isolated from the Mexican tree Hintonia latiflora, Sporomiella minimoides (FQNP587)16. Figure 2 shows the chemical structure of those compounds.
For visualization of the chemical space of the datasets compared in this work, we computed six physicochemical properties of pharmaceutical relevance. As described in the Methods section, we computed the correlation matrix between them (Extended data, Table S1 in Supplementary File 2) and since all descriptors were highly correlated (correlation coefficient > 0.60), with the exception of SlogP, we selected this and its lowest correlated property, AMW, as the axes to project the libraries. We also carried out a principal component analysis over the scaled properties. As expected from the correlation matrix, the first principal component was associated with all properties but SlogP and in the opposite way for the second principal component, with explained variances of 69% and 19% respectively. Therefore, the projection of the data over those two components looks quite similar to the selected properties (Slogp and AMW) (Extended data, Figure S1 in Supplementary File 2), with the difference that the principal components do not have the physical interpretation that properties do, e.g. the orally available drugs space as described by Lipinski17. Figure 3 shows a visual representation of the chemical space of the three datasets analyzed in this work. For this plot, in order to better illustrate the unique compounds present in BIOFACQUIM, those compounds belonging to two libraries were assigned to a single one: ChEMBL if they belong to this dataset and NPs if they belong to this dataset but not to ChEMBL. Compounds shared among the three libraries were assigned to a different category. This figure shows that ChEMBL is the most widespread dataset in terms of physicochemical properties and that NPs, BIOFACQUIM, as well as the shared compounds among the three datasets, are mostly contained within this space. It could be noted that a few exceptions from BIOFACQUIM cover a small part of the space currently unexplored by ChEMBL and it is also remarkable that contrary to ChEMBL and NPs, most BIOFACQUIM compounds are part of the orally available drugs space. Different panels showing all different overlaps between datasets can be found in Extended data (Figure S2 in Supplementary File 2).
In order to compare the chemical diversity of the current version of BIOFACQUIM with the previous one and the two datasets selected as reference, we employed a CD plot Figure 4 shows the plot comparing the diversity of all datasets considering four different criteria: scaffolds in the y-axis, molecular fingerprints in the x-axis, physicochemical properties as the filling of the data points in a continuous color scale, and number of compounds as the data points size. This comparison shows the relatively small size of BIOFACQUIM in comparison with the reference datasets. As compared to the previous release of BIOFACQUIM, the current version has increased its diversity in terms of scaffolds and fingerprints but decreased in terms of physicochemical properties. Also, it is shown that its diversity in terms of molecular fingerprints and physicochemical properties, although not the greatest ones of the three datasets, are on the order of those for NPs, contrary to scaffolds, in which is the most diverse.
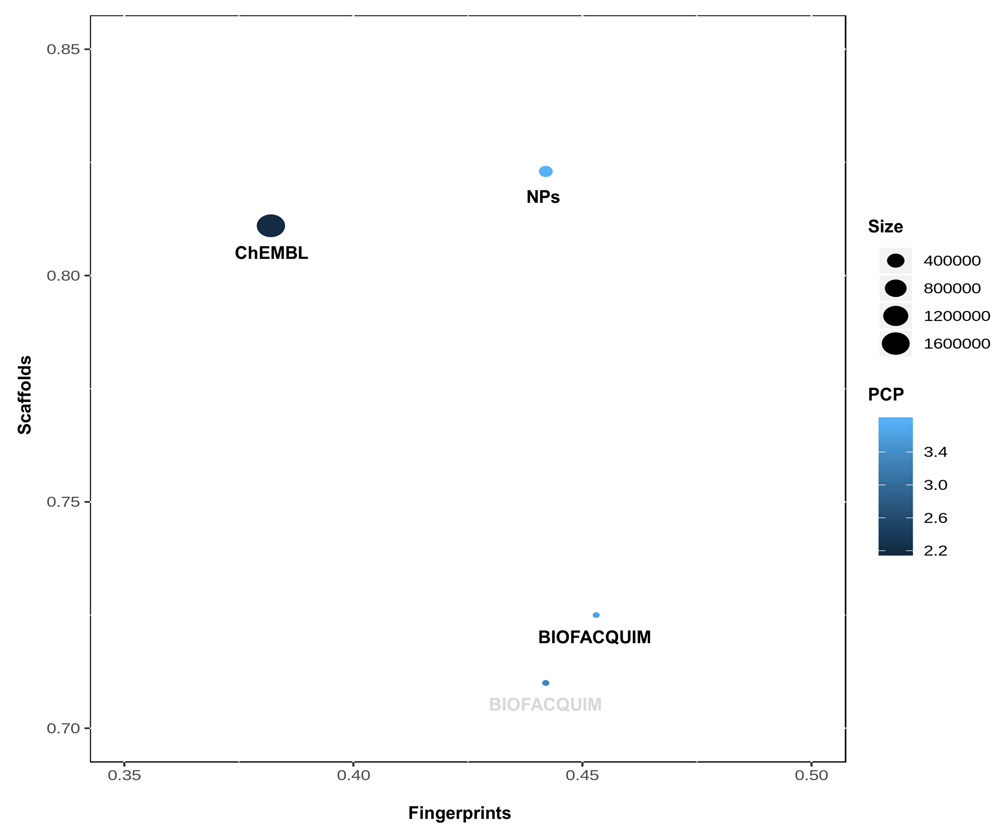
The molecular fingerprint diversity of each data set is represented on the x-axis and was defined as the median Tanimoto coefficient of MACCS keys (166-bits) fingerprint. The scaffold diversity of each database is represented on the y-axis and was defined as the area under the corresponding cyclic system retrieval curve. The diversity based on physicochemical properties (PCP) was defined as the mean euclidean distance of six scaled physicochemical properties (SlogP, TPSA, AMW, RB, HBD, and HBA) and is shown as the filling of the data points using a continuous color scale. The number of compounds is represented by the size of the data points.
The current version of BIOFACQUIM involved the addition of 148 natural products. This was reflected in a diversity increase based on both scaffolds and molecular fingerprints. It was shown that in terms of diversity and structural content overlap, BIOFACQUIM is more similar to the assembled set of natural products than to the set of biologically tested compounds. The herein reported chemoinformatic study revealed that most of the compounds contained in BIOFACQUIM are focused in the orally active drugs space in terms of physicochemical properties. Interestingly, despite the fact of its relative small size, there were identified a significant number of compounds, scaffolds and functional groups (81, 28 and 5, respectively) that were not present in the two large sets used as reference, showing that curated databases of natural products, such as BIOFACQUIM, can serve as a starting point for the study and increase of the biologically relevant chemical space.
Figshare: BIOFACQUIM_V2. http://doi.org/10.6084/m9.figshare.11312702
This file contains the chemical structures of 531 compounds in SDF format, alongside ID number, compound name, simplified molecular input line entry system, literature reference, kingdom, genus, species, geographical location and biological activity.
Underlying data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
Figshare: Supporting information for "Functional group and diversity analysis of BIOFACQUIM: A Mexican natural product database". http://doi.org/10.6084/m9.figshare.11312735
This project contains the following extended data:
Supplementary File 1. File with summary results of the functional group analysis.
Supplementary File 2. This file contains the following supporting tables and figures:
Table S1. Correlation matrix for the six PCP computed for BIOFACQUIM and the two reference datasets.
Figure S1. Visual representation of the chemical space covered by the three compound libraries generated by principal component analysis.
Figure S2. Visual representation of the chemical space covered by the possible overlaps between libraries.
Extended data are available under the terms of the Creative Commons Attribution 4.0 International License (CC BY 4.0).
We thank Roberto Hernández for his valuable contribution toward the update of BIOFACQUIM.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Natural product chemistry, Drug Discovery.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Computational Chemistry, Cheminformatics, Drug Design
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: cheminformatics, natural products research
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 08 Jun 20 |
read | ||
|
Version 1 10 Dec 19 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)