Keywords
Patient-Derived Xenografts, Triple negative breast cancer, Luminal B breast cancer, Interventional research
This article is included in the Oncology gateway.
Patient-Derived Xenografts, Triple negative breast cancer, Luminal B breast cancer, Interventional research
We added relevant literature in the introduction section. We also cited five new references: Bruna et al. (2016) (PMID: 27641504), DeRose et al. (2011) (PMID: 22019887), Sachs et al. (2018) (PMID: 29224780), Rosenbluth et al. (2020) (PMID: 32249764) Ben-David et al. (2017) (PMID: 28991255). Some details have been added about the PDXs commercialization and production
See the authors' detailed response to the review by Fares Al-Ejeh
See the authors' detailed response to the review by Jessica Finlay-Schultz
Breast cancer can be classified into different molecular subtypes according to gene expression: luminal A (Hormone Receptor positive (HR+)/human epidermal growth factor receptor 2 negative (HER2-), with high HR expression and low levels of the protein Ki-67), luminal B (HR+ either HER2+ or HER2- with high levels of Ki-67 and with low HR expression), HER2-enriched (amplification of ERBB2 gene, regardless of HR status) and triple-negative (HR-/HER2-). Luminal B and triple-negative tumours account respectively for approximately 20% and 10–15% of all breast cancers1–3. Although rarer than luminal A breast cancers, luminal B and triple-negative tumours are often high-grade tumours with a poorer prognosis1.
The establishment of patient derived xenografts (PDX) could be useful to discover new treatments and strategies needed in the fight against these subtypes of breast cancer. PDX are derived from tumour tissue in which the tumour architecture and the proportion of cancer and stromal cells are both maintained: advantages not found in cell lines. Therefore, PDX effectively model intra- and inter-tumoural heterogeneity4,5. It is important to note that PDXs have been shown to undergo mouse-specific tumour evolution6.
Thus, PDX are used to answer questions such as the contribution of tumour heterogeneity to therapeutic response, patterns of tumour progression during metastatic progression and mechanisms of treatment resistance7,8.
Most of the available PDX or organoids, which have been xenografted into mice and recapitulate the primary tumours, are generated from primary breast cancer tissue5,9–11. In contrast, generated PDX from metastatic tumours remain rarer and may allow the identification of eventual molecular therapeutic targets in metastatic setting.
It is also necessary to have PDX models from primary breast lesions that are resistant to neoadjuvant therapies. Recent prospective studies show that PDX can be obtained from neoadjuvant breast tumours and demonstrate the feasibility of tumour sequencing in these situations12,13. Breast cancer patients with residual disease after neoadjuvant chemotherapy (NAC) have an increased risk of recurrence. Similarly, high-grade breast tumours treated by primary surgery are very rare, poorly known, and aggressive.
The production of PDX from post-NAC residual breast tumours or from high-grade breast tumours will provide data on the molecular characteristics of these tumours with a high risk of recurrence.
In this study, we want to establish PDX from tumour samples of patients with triple-negative and luminal B breast cancers in neoadjuvant, adjuvant or metastatic settings. In addition, to verify whether or not the PDX obtained is consistent with the original tumour, we will study the tumour exomes of both the PDX and the original tumour. The study of the patients’ constitutional exome will serve as the basis for this comparison and is an essential element in the overall somatic analysis.
This is a single-centre prospective trial designed to establish xenografts from surgical specimens of patients with triple negative or luminal B breast cancer in neoadjuvant, adjuvant or metastatic setting. Patient enrolment is expected to take 3 years: 85 patients will be enrolled and followed during 28 months.
Study design is presented in Figure 1. The management of patients in the study may vary depending on the setting: neo-adjuvant, adjuvant or metastatic. In all the settings and as a part of their medical follow-up, patients will go through a pre-operative biological assessment. For patients in metastatic setting, this assessment will be performed before surgical excision of the metastases. Blood samples will be used to perform sequencing of the patient's constitutional exome. At the end of the surgery, one sample of the surgical specimen will be taken to generate PDX and another one to sequence the patient's tumour exome.
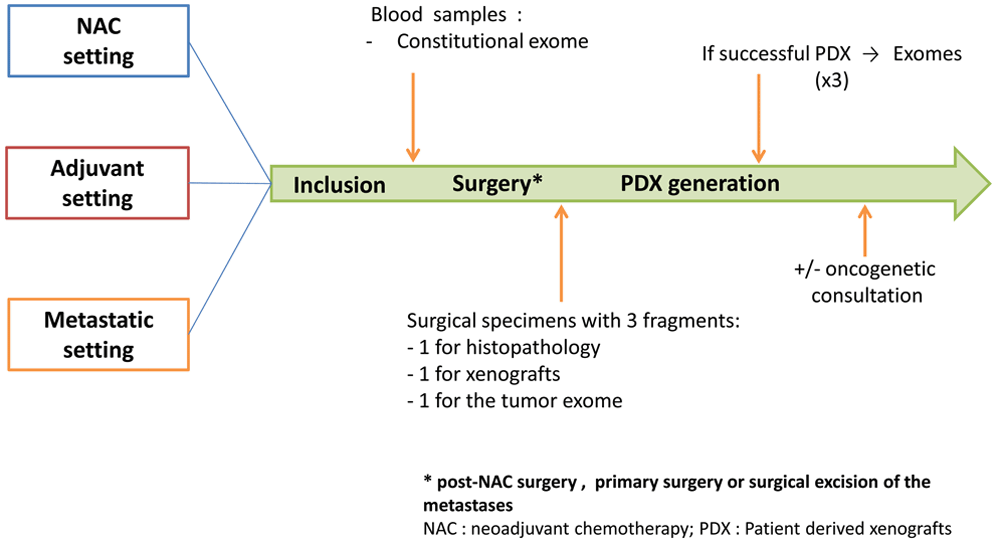
Adult women with triple negative or luminal B breast cancer in neoadjuvant, adjuvant or metastatic setting will be included. In all the settings, patients will go through a pre-operative biological assessment. Blood samples will be used to perform sequencing of the patient's constitutional exome. At the end of the surgery (post-NAC surgery, primary surgery or surgical excision of the metastases), a sample of the surgical specimen will be taken to generate patient-derived xenografts (PDX) and another one to sequence the patient's tumour exome.
Participants can withdraw at any time. Data obtained will be retained with consent, and any reasons given for withdrawal will be recorded.
The patient-derived tumour xenograft platform called XenTech will generate the PDX. All PDX will be established as approved by the ethical authorisation #16569: « Développement d’une collection de modèles de tumeurs humaines greffés sur souris (PDX) ». The different steps of the development of an established xenograft model are summarized in Figure 2. Overall, fresh surgical tissue will be cut into small fragments and grafted into the inter-scapular region or into the renal capsule of 6- to 13-week-old female immunodeficient or severe-combined immuno-deficiency (SCID) or non-obese diabetic SCID mice. The inter-scapular site allows easy access to a well vascularized region with tissue that in female mice is in continuity with the mammary gland. Although the use of extracellular matrix tends to increase xenograft take rates, tumor fragments will be implanted without any matrix. Mice should weigh 18g at 6 months. At the onset of tumour growth, a latency period of 1 to 9 months is expected. The mouse generation with the patient derived graft will be called F0 and the following generations will be numbered F1, F2, and F3. When the tumour volume reaches the ethical limit (10% of the total weight of the mouse), the tumour is removed and fragmented as follows: a part is grafted to a new set of mice; a second part is fixed in formalin and embedded in paraffin for histological studies; a third part is frozen in liquid nitrogen for molecular studies; and a last part is frozen in a 10% DMSO solution to generate a viable tissue stock. These steps are repeated for the F1, F2 and F3 generation. A xenograft will be considered as well-established in the third generation (F3). When it is necessary, the animal will be sacrificed by cervical dislocation. For the duration of the study, mice will be housed, by a maximum of 6, in individually ventilated cages. They will be housed in a light-dark cycle with temperature and hygrometry control. The mice will have throughout the duration of the project a complete diet and drinking water. For the ER+ PDX models, mice will receive 17β-estradiol supplementation in the drinking water, useful for the establishment of these models14. No estrogen pellets will be implanted.

Fresh surgical tissue will be grafted into the inter-scapular region or into the renal capsule of immunodeficient or severe-combined immuno-deficiency mice. When the tumour volume reaches the ethical limit, the tumour is removed and fragmented as follows: a part is grafted to a new set of mice; a second part is fixed in formalin and embedded in paraffin for histological studies; a third part is frozen in liquid nitrogen for molecular studies; and a last part is frozen in a 10% DMSO solution to generate a viable tissue stock. These steps are repeated for the F1, F2 and F3 generation. A xenograft will be considered as well-established in the third generation (F3).
Inclusion and exclusion criteria are presented in Table 1. Briefly, adult women (18 years or older) with triple-negative or luminal B breast cancer in neoadjuvant, adjuvant or metastatic setting will be included.
Eligible patients will be offered the opportunity to participate in the study by their oncologist or their surgeon. Patients who agree to participate in this study will provide written informed consents (clinical consent and genetic consent) for enrolment. XenTech is allowed to commercialize the PDX generated in this project. The patients will be made aware of that during consent.
This feasibility study aims to obtain xenografts from tumours that are either rare or tumours that are resistant to treatment and therefore difficult to establish. We consider that a minimum of five successful grafts would meet this objective. Under these conditions, knowing the graft success rate specific to each histological type and the proportion of luminal B (2/3) and triple-negative breast cancers (1/3), we calculate the number of subjects required so that the lower bound of the 95% confidence interval (CI) of the success rate multiplied by the number of subjects is ≥5.
To obtain a xenograft, the average rate is 30% from triple-negative tumours and 10% from luminal B tumours. Taking these data into account, we will include 2/3 luminal B tumours and 1/3 triple negative tumours: the expected rate of successful xenografts will be approximately 17%. The number of patients needed to be included is therefore at least 65: lower bound of the CI-95% of 17% for 65 subjects = 8% or 5 patients (8%×65=5.2).
Since for some patients the tumour will be of sufficient size for imaging, but of insufficient size in the operating room, we consider that this number should be increased by 30%, to a total number of 85 patients.
The primary objective of the XENOBREAST trial is to establish xenografts from tumour samples of patients with triple negative or luminal B breast cancer in neoadjuvant, adjuvant or metastatic setting. Furthermore, the study aims to investigate histomolecular characteristics of the established PDX and to compare these characteristics with the initial histomolecular profile of the collected tumours.
Data collected are the patient's age (month and year of birth), pathology, treatments received, response to treatments, date and nature of surgery (primary tumour, metastasis), data concerning exomes (constitutional and tumour), as well as histomolecular profiles of the initial tumour and of the different xenograft generations. To define these histomolecular profiles we will quantify the expression of the oestrogen, progesterone and androgen receptors by immunohistochemistry, the amplification status of the ERBB2 gene, and the fraction of tumour cells expressing Ki67. Finally, tumours will be classified into molecular classes according to the above-mentioned data: luminal A, luminal B, HER2-enriched or triple negative.
Data collected and transmitted to the sponsor of the study by the investigators will be pseudonymized. Study data will not contain any names or other personal identifiers such as addresses. Patients included in the trial will be identified by a code specific to this trial. The investigator will have access to the correspondence table between the patient's last name, first name, date of birth and the code assigned in the trial.
Primary analysis. The main outcome of this feasibility trial is the number of successful xenografts obtained, a xenograft being considered successful when it reaches the F3 generation and its molecular subtype defined by immunohistochemistry has remained identical to the original tumour. The percentage of tumours yielding a successful xenograft will also be calculated, along with a 95% confidence interval. The feasibility will be considered acceptable if at least five successful transplants are obtained.
Secondary analysis. A diversity analysis of genomic data from the tumour exomes and the constitutional exome will be performed. Differential analyses using bioinformatics tools adapted to these data could be envisaged if the number of patients allows it. We will describe in detail the collected tumours (description of the population, histomolecular profile, anatomopathological data, etc.). Qualitative characteristics will be described using their number and frequency, and quantitative characteristics (age at diagnosis, tumour size, etc.) using standard distribution parameters: mean, median, standard deviation, extremes, normality.
The characteristics of the tumours will also be described according to whether or not a xenograft was successful. These characteristics will also be compared, if the sample sizes allow it, using Fisher’s exact test, and Welch's t-test or the non-parametric Mann-Whitney U-test if needed. All tests will be two-sided and the statistical significance threshold will be generally set at 0.05 except in case of differential analyses on the exome data where multiple testing corrections will be applied.
The XENOBREAST trial has been approved by an ethics committee (Sud Méditerranée IV – Montpellier) on April 2020 (Reference: 20 03 02 and ID-RCB number: 2020-A00398-31). It is conducted notably in accordance with the Declaration of Helsinki and General Data Protection Regulation (GDPR).
Study data and finding will be published in peer-reviewed medical journals. We plan to present the study and all data at national congresses and conferences.
The XENOBREAST study a feasibility pilot trial that will allow us to estimate the success rate of xenografts and to estimate PDX drift by comparing the histomolecular profile of the PDX to that of the tumour. In perspective, the generation of PDX from rare and chemo-resistant tumours would allow for testing new treatments before their administration in vivo. In the long term, the establishment of PDX from primary mammary tumours or after neoadjuvant treatment would allow a better understanding of the therapeutic response. Moreover, it could be a great model to explore tumour evolution patterns during metastatic progression and to observe tumour resistance mechanisms in non-metastatic tumours.
No data are associated with this article.
The authors thank the patient-derived xenografts platform XenTech for their help in the study design and in the PDX xenografts establishment. The authors also thank the Laboratoire d’Oncogénétique of the Jean PERRIN centre of Clermont-Ferrand.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Breast cancer research and translational in vitro and in vivo models, including PDXs and PDCLs. My area of research is cancer signalling and identification of biomarkers and therapeutic targets from clinical samples to investigate their role and clinical potential in preclinical models.
Is the rationale for, and objectives of, the study clearly described?
Yes
Is the study design appropriate for the research question?
Yes
Are sufficient details of the methods provided to allow replication by others?
Partly
Are the datasets clearly presented in a useable and accessible format?
Not applicable
References
1. Bruna A, Rueda O, Greenwood W, Batra A, et al.: A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell. 2016; 167 (1): 260-274.e22 Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Breast cancer research and translational in vitro and in vivo models, including PDXs and PDCLs. My area of research is cancer signalling and identification of biomarkers and therapeutic targets from clinical samples to investigate their role and clinical potential in preclinical models.
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: My expertise is in the establishment and propagation of breast cancer PDX models, primarily from hormone receptor positive tumors. My area of research includes estrogen and progesterone receptor signaling, crosstalk, and transcriptional regulation in breast cancer models, including PDX and cell lines derived from primary tissue and PDX models.
Is the rationale for, and objectives of, the study clearly described?
Yes
Is the study design appropriate for the research question?
Yes
Are sufficient details of the methods provided to allow replication by others?
No
Are the datasets clearly presented in a useable and accessible format?
Not applicable
References
1. Kabos P, Finlay-Schultz J, Li C, Kline E, et al.: Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures.Breast Cancer Res Treat. 2012; 135 (2): 415-32 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: My expertise is in the establishment and propagation of breast cancer PDX models, primarily from hormone receptor positive tumors. My area of research includes estrogen and progesterone receptor signaling, crosstalk, and transcriptional regulation in breast cancer models, including PDX and cell lines derived from primary tissue and PDX models.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 3 (revision) 21 Jun 21 |
read | |
|
Version 2 (revision) 10 Mar 21 |
read | read |
|
Version 1 09 Oct 20 |
read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)