Keywords
NEFH-tTA, frontotemporal dementia, FTD, ALS, targeted locus amplification, genotyping, mouse
This article is included in the Genomics and Genetics gateway.
NEFH-tTA, frontotemporal dementia, FTD, ALS, targeted locus amplification, genotyping, mouse
Following constructive feedback from 3 reviewers, we have made some minor changes to this manuscript. The main changes are detailed below:
- The phrase "copy number" has been changed to "zygosity" in the title
- Fig 3 which was a pie chart has been removed and replaced with Table 5
- A FASTA file showing the sequence of the transgene has been Underlying Data repository, which can be found here https://figshare.com/articles/dataset/FASTA_reference_file_for_the_NEFH-tTA_transgene/16847170
See the authors' detailed response to the review by Keiichi Ishihara
See the authors' detailed response to the review by Virginia Lee and Mandana Hunter
See the authors' detailed response to the review by Sandy S. Pineda
NEFH, neurofilament heavy polypeptide promoter; TG, transgene; tTA, tetracycline-controlled transactivator protein; TRE, tetracycline response element; TLA, targeted locus amplification.
The NEFH-tTA mouse has the human neurofilament heavy polypeptide promoter directing tetracycline-controlled transactivator protein (tTA) expression to large-calibre axons of the brain and spinal cord, allowing tissue-specific and doxycycline-suppressible expression of a target gene1. The line was generated via pronuclear injection of a plasmid with the human NEFH promoter isolated from BAC clone (CHORI: RP11-91J21) to drive expression of the tetracycline transactivator gene, which randomly integrated into the genome1. While a genotyping protocol is available to distinguish between wild-type and transgenic animals1, ideally a colony of homozygous animals would be maintained for experimental breeding with another transgenic line with the gene of interest under the control of a tetracycline response element (TRE). All offspring of these breeds would therefore be NEFH-tTA+/- (which has been shown to be a sufficient driver of expression1); this would allow use of littermate controls and reduce required breeding numbers as encouraged by the ARRIVE guidelines2. In order to address this, we have identified the insertion site of the NEFH-tTA transgene, designed a novel PCR genotyping strategy, and demonstrated its reliability in a colony of NEFH-tTA mice.
All animal work was approved following local ethical review by the University of Manchester Animal Welfare and Ethical Review Board and performed under Home Office project license 70/8903 and in accordance with the Home Office (Animals) Scientific Procedures Act (1986). All efforts were made to ameliorate harm to animals; mice were housed and maintained according to standard practices in the University of Manchester Biological Services Facility (detailed below) and no licenced procedures were performed.
B6;C3-Tg(NEFH-tTA)8Vle/J mice (JAX stock #025397) were obtained from Jackson Laboratories (Bar Harbor, ME, USA) and bred with C57BL6/J mice to produce a colony of hemizygous NEFH-tTA animals (13 transgenic mice were crossed with 12 wild-type in total, as this number was sufficient to create a new colony of backcrossed mice). All animal work was carried out in the University of Manchester Biological Services Facility. Mice had free access to food and water and were housed under light-humidity- and temperature-controlled conditions: ambient temperatures of 21°C (±2°C), humidity of 40–50%, 12 h light cycle, ad libitum access to water and standard rodent chow. Animals were housed up to five per cage with environmental enrichment. For breeding, mice were housed as either breeding pairs or trios of two female and one male, and pups housed with parents until weaning (approximately four weeks post-birth).
At weaning, animals were restrained by scruffing and ear biopsies were taken using a standard ear puncher and genotyped as described1. Briefly, tissue was disrupted in proteinase K for one hour at 57°C; reactions containing 3μL of diluted DNA, 10μL PrimeSTAR HS Premix (Takara, Clontech #R040A) and 0.5μM each of forward and reverse transgene primers and internal positive control primers (Primers 1–4, Table 1) in a total volume of 20μL were amplified in a thermal cycler (ThermoFisher SimpliAmpTM A24812; Table 2). PCR products were visualised on a 2% agarose gel with HyperLadder IV (Bioline) to determine product size.
| No. | Sequence | |
|---|---|---|
| 1 | CTC GCG CAC CTG CTG AAT | Transgene allele detection1 |
| 2 | CAG TAC AGG GTA GGC TGC TC | Transgene allele detection |
| 3 | CTA GGC CAC AGA ATT GAA AGA TCT | Internal control1 |
| 4 | GTA GGT GGA AAT TCT AGC ATC ATC C | Internal control |
| 5 | TCATGGAGACACCAATGACG | Wild-type detection |
| 6 | GGAGGAATTCATGTGCCACT | Wild-type detection |
A combination of targeted locus amplification (TLA) and next-generation sequencing was used to identify the insertion point of the NEFH-tTA transgene (the FASTA of the transgene sequence can be viewed in the Underlying data). This was performed by Cergentis B.V (Utrecht, The Netherlands). One mouse spleen was dissected from a six-week old NEFH-tTA+/- mouse (culled by Schedule 1 CO2 inhalation) and cells isolated for TLA sample prep. Spleen tissue was disrupted by gently pushing through a 40µm mesh and cells collected in PBS with 10% foetal calf serum (Gibco #10500064). Cells were pelleted by centrifugation at 250g for ten minutes at room temperature, resuspended in lysis buffer (0.15M NH4CL, 0.01M KHCO3, 0.0002M EDTA) and incubated at room temperature for five minutes before additional centrifugation at 250g for ten minutes. Cells were resuspended in PBS with 10% foetal calf serum (Gibco #10500064), stored at -80°C and shipped to Cergentis on dry ice.
Two primer sets (Table 3) were used in individual TLA amplifications. PCR products were purified and library prepped using the Illumina NexteraXT protocol and sequenced on an Illumina sequencer. Reads were mapped using BWA-SW, which is a Smith-Waterman alignment tool. This allows partial mapping, which is optimally suited for identifying break-spanning reads. The mouse genome version mm10 (GenBank assembly accession: GCA_000001635.2) was used for mapping.
Hemizygous mice were bred together (total 25 animals; the number of breeding animals was chosen based on expected number of homozygous offspring and numbers required to set up a large colony to breed with a different transgenic line3) and ear biopsies taken from all offspring at weaning (n=50) as described above. Primers were designed using Primer3 (web version 4.1.0) to produce a product spanning the insertion site on chromosome 12 (Primers 5-6, Table 1) and PCR reactions were set up as described under Maintenance of transgenic mice. REDExtract-N-Amp™ 2X PCR Reaction Mix (Sigma-Aldrich, #R4775) was used instead of PrimeStar.
Outcome vs Expected for genotype was performed using GraphPad Prism version 8.00 for Mac (GraphPad Software, La Jolla California USA, www.graphpad.com).
Using TLA highest coverage is observed on the sequences directly surrounding the location of the primer set. Here, high coverage is observed on chromosome 12, outlined in red in Figure 1A, indicating the transgene (TG) has integrated in chromosome 124.
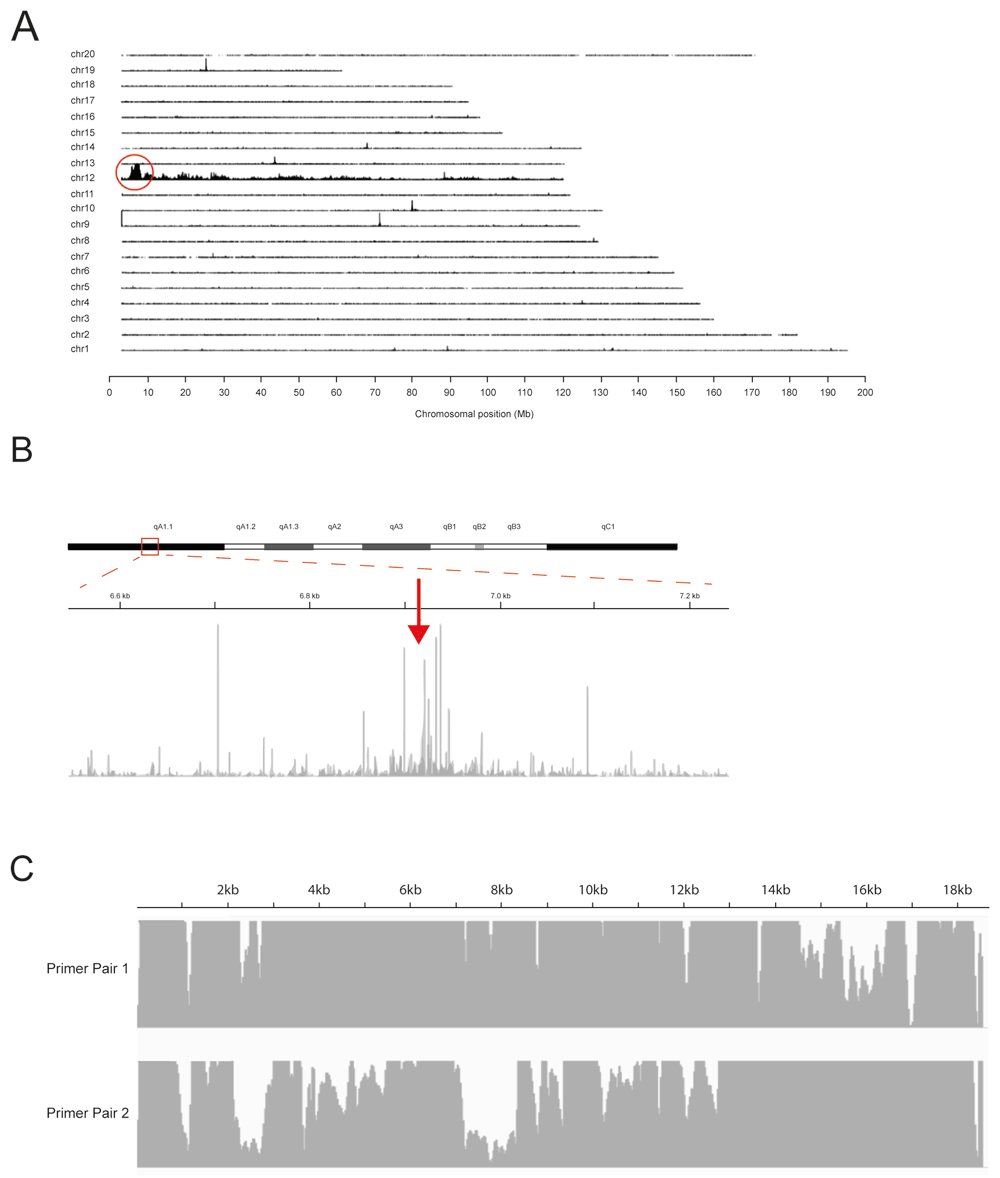
(A) Targeted locus amplification (TLA) sequence coverage across the mouse genome using primer set 1. The chromosomes are indicated on the y-axis, the chromosomal position on the x-axis. Similar results were obtained with primer set 2 (see Underlying data4). (B) TLA sequence coverage across mouse chromosome 12 with primer pair 1. The arrow points towards the integration site. (C) TLA sequence coverage across the TG with primer pair 1 (top) and 2 (bottom).
From locus-wide coverage (Figure 1B) we were able to identify the junction sites between the TG and the mouse genomic sequence, and it was concluded that the TG has integrated in mouse chr12:6917896-chr12:6917912. Reads marking the TG integration are identified in Table 4. The 11 bases between chr12:6917896-chr12:6917912 have been deleted following the integration event. According to the reference sequence (mm10) there are no genes annotated in this region. Complete coverage is observed across the whole TG sequence from TG:68-18543 (Figure 1C). At the edges of the coverage, fusion reads have been found, connecting these ends, but the TG: 1 -68 and TG 18544 – end regions are not integrated into the mouse genome (Table 4). These regions contained sequences for bacterial DNA replication and therefore are not required for transgenic expression in the mouse.
Homologous sequences are shown in purple.
Next to the head-to-tail fusion as reported above, four other TG-TG fusions were found within the TG sequence (Table 4). While an exact copy number cannot be determined using TLA, an estimation can be made based on the number of integration sites, number of fusion reads and the ratio of coverage on the TG and genome integration site. Here, one integration site was found and a total of five TG-TG fusions were found. The coverage at the TG was significantly higher compared to the genome. These facts together suggest that >5 copies of the TG may have integrated.
We designed primers to amplify a wild-type PCR product of 488bp from chromosome 12, which would be disrupted by insertion of the transgenic allele (Figure 2A). A multiplex PCR reaction including the original primers to the transgene1 (Table 1) was carried out on samples from a hemizygous intercross and proved able to distinguish between wild-type (488bp band only), hemizygous (both bands) and homozygous (150bp band only) (Figure 2B)4.
After genotyping 50 offspring of hemizygous intercrosses we observed no significant difference in the number of Nefh-tTA+/+ mice compared to the expected ratio (Table 5, chi-square=2.64, p<0.26714), confirming our novel genotyping assay performs as expected. Homozygous mice grew to adulthood and did not exhibit any overt phenotype.
A chi-square test was performed (using GraphPad Prism version 7.04) to confirm that the observed numbers were not significantly different to expected, indicating that our genotyping method works as predicted.
The NEFH-tTA transgenic mouse line is a useful tool for studying a wide range of diseases including frontotemporal dementia and motor neurone disease, as well as other neurodevelopmental, neuromuscular or neurodegenerative disorders. Here we have shown that the transgene has inserted into chromosome 12. Furthermore, we have designed and utilised a novel genotyping assay to distinguish between hemizygous and homozygous mice, involving a simple PCR assay. This is easily adaptable to a laboratory-specific protocol or machine and will allow researchers using this line to refine their breeding strategies and reduce the number of animals that cannot be used in experiments.
Figshare: “A novel genotyping method to determine zygosity in a mouse line commonly used for inducible transgene expression in brain and spinal cord – Raw data/analysis” DOI: https://doi.org/10.6084/m9.figshare.12982220.v14.
This project contains the following underlying data:
Figshare: FASTA reference file for the NEFH-tTA transgene, https://doi.org/10.6084/m9.figshare.16847170.v15
This project contains the following underlying data:
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
We would like to thank the University of Manchester Biological Services Facility for animal care.
Prof Stuart Pickering-Brown, the last author listed on this article, has sadly passed away since submission of the original version. This publication is in his memory.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Neuroscience, Pathological biochemistry
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Neuroscience.
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Neuroscience, Pathological biochemistry
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 10 Feb 22 |
read | ||
|
Version 1 15 Oct 20 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)