Keywords
Extracellular vesicles, exosomes, survivin/BIRC5, long-noncoding RNA, CELLine bioreactor, HOTAIR
This article is included in the Cell & Molecular Biology gateway.
Extracellular vesicles, exosomes, survivin/BIRC5, long-noncoding RNA, CELLine bioreactor, HOTAIR
We have replaced the previous TEM image with an improved TEM image consisting of multiple EVs in Figure 1B.
To read any peer review reports and author responses for this article, follow the "read" links in the Open Peer Review table.
Interactions between tumour and stromal cells sculpt the tumour microenvironment and contribute to cancer malignancy, metastasis and immune evasion. Extracellular vesicles (EVs)1 mediate one of the key intercellular interactions by shuttling biomolecules in micro and nanoscale lipid-enclosed packages. EVs have been associated in many studies with resistance of cancer to chemo or radio therapies2.
EVs contain cargo specific to their parental cell, are very stable, and circulate in blood and other bodily fluids. These properties make EVs prime candidates for cancer detection in liquid biopsies3, either alone or combined with the detection of circulating tumour DNA (ctDNA) or circulating tumour cells (CTCs)4. Upregulation of RNA transcripts including long-noncoding RNA (lncRNA) offers a means for distinguishing EVs originating from tumour and non-tumour cells. LncRNAs are greater than 200 nucleotide-long transcripts constituting two thirds of the transcriptome and appear to play a critical role in carcinogenesis of many cancers including breast malignancies5–11, and constitute an understudied class of EV cargo12,13. HER2-positive breast cancers were reported to differentially express over 1,300 unique lncRNAs compared to non-tumour breast tissue14,15. Some of the HER2-associated lncRNAs identified to date include ZFAS1 which was found to associate with ribosomes in breast cancer cells16,17; HOX transcript antisense intergenic RNA (HOTAIR) which is involved in regulation of chromatin states and targets genes related to tumour metastasis and correlates with poor prognosis18,19; and AGAP2 antisense RNA 1 (AGAP2-AS1) which promotes resistance of breast cancer cells to trastuzumab through EV signalling20. Some mRNAs specific to or upregulated in breast cancer cells may also serve as HER2-EV biomarkers, including EpCAM which affects intercellular adhesion and is relevant to tumour progression21; BIRC5 (baculoviral IAP repeat-containing protein 5, the gene that encodes the survivin protein and a member of Inhibitor of apoptosis family) which is involved in regulation of mitosis and apoptotic inhibition22; and YBX1 (the Y-box binding protein) which is an oncogenic transcription and translation regulator that regulates cell invasion and migration23. The overexpression of these mRNAs (EpCAM, BIRC5 and YBXI) were all associated with poor outcomes for breast cancer patients24. RNAs represent promising EV-associated biomarkers but difficulties in producing sufficient amounts of pure cancer associated EVs complicate validation of RNA presence in EVs.
Here, we present a simple solution for obtaining high quantities of cancer-associated EVs by culturing the HER2-positive breast cancer cell line BT-474 in a CELLine AD 1000 two-chamber bioreactor flask. The CELLine bioreactor system mimics physiological growth conditions by allowing three dimensional (3D) cell growth on a fibre-mimetic surface, resulting in increases in cell number and EV production25. This strategy allowed us to obtain sufficient EV yields to demonstrate that tumour cells release EVs associated with several potential breast cancer biomarkers.
To prevent bovine EVs present in foetal calf serum (FCS) from contaminating the cancer-specific EVs, we cultured BT-474 cells from ATCC (ATCC® HTB-20™) (seeded at 4.5 × 108 cells/mL) in 15 mL Advanced DMEM/F-12 medium (Gibco, ThermoFisher Scientific, Waltham, USA) supplemented with 2% CDM-HD serum replacement (FiberCell Systems, New Market, USA) in the lower cell chamber of a CELLine AD 1000 bioreactor flask (Argos, Elgin, USA). The same media (150 mL) was used in the upper media chamber but supplemented with 2% FCS (Figure 1A). The dialysis membrane that separates the cell and media compartments allows FCS-specific nutrients <10 kDa but not EVs to pass through and nourish the cells. Every three to four days, the 15 mL of conditioned medium from the cell chamber was harvested for EV isolation, and the media from the upper chamber was replaced.

(A) experimental procedure employed for extracellular vesicle (EV) production, isolation, and purification; (B) transmission electron microscopy image of a small EV; (C,D) hydrodynamic diameter distribution profiles of isolated large and small EVs measured by nanoparticle tracking analysis (NTA) wherein red vertical lines and blue numbers denote standard deviation and particle diameters at specific peaks, respectively; (E) EV concentration (empty squares) determined by NTA, and protein levels (filled squares) determined by BCA assay of fractions acquired during separation on a qEV Original size exclusion chromatography (SEC) column; and (F) immunoblot with antibodies specific for HER2, EpCAM, α-tubulin and TSG101 proteins. Tetraspanin TSG101 is a loading control expected to be present in both EVs and cells. The α-tubulin should be present only in the cell lysates but not in purified EVs. MDA-MB-231 cell lysate serves as the negative control for HER2 and EpCAM proteins. Representative images/data from three independent experiments were shown in B–F.
EVs were isolated using differential centrifugation and size exclusion chromatography (SEC) as outlined in Figure 1. Conditioned medium (15 mL) was first centrifuged at 2,000 x g for 10 min to remove large debris, 10,000 x g for 30 min to isolate large EVs, and 100,000 x g for 70 min to isolate small EVs (Figure 1A). The resulting small EV suspension (in 500 µL PBS) was loaded onto a 35 nm qEV original size exclusion column (Izon, Christchurch, New Zealand), and fractions 7 through 24 were collected using an automated fraction collector (500 µL per fraction). BCA protein quantitation assay (Cat # 23225, Pierce, ThermoFisher Scientific, Waltham, USA) and Nanosight NS300 nanoparticle tracking analysis (NTA; Malvern Panalytical, Malvern, UK) were performed to quantitate protein and particle concentrations in each fraction, respectively. EV concentrations and size distributions were quantified on NTA by recording three 30 seconds videos under low flow conditions, with large EVs diluted at 1:100 in PBS and small EVs diluted at 1:500 in PBS. Small EV-rich fractions (7–11) were pooled, quantified again using NTA and BCA, and concentrated by ultracentrifugation (Avanti, Beckman Coulter, Brea, USA) at 100,000 x g for 70 min.
Negative staining TEM of pooled EV fractions was conducted by adsorption onto Formvar-coated copper grids (Electron Microscopy Sciences, Hatfield, USA) for 2 min, then treated with 2% uranyl acetate for 2 min. Grids were then visualised on a Tecnai G2 Spirit TWIN (FEI, Hillsboro, OR, USA) transmission electron microscope at 120 kV accelerating voltage and images were captured using a Morada digital camera (SIS GmbH, Munster, Germany).
This procedure was carried out as described previously26. Breast cancer cell lines were grown to log-phase, washed twice with ice-cold PBS, and lysed in an sodium dodecyl sulphate (SDS) lysis buffer [60 mM Tris-HCl (pH 6.8 at 25°C), 2% (w/v) SDS, 10% glycerol]. Proteins (25 μg) were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to PVDF membranes. Membranes were subsequently immunoblotted with antibodies recognising human HER2 (mouse monoclonal, anti-Neu, Santa Cruz, Cat # sc-33684, RRID:AB_627996), human EpCAM (rabbit monoclonal, AbCAM, Cat # ab223582, RRID:AB_2762366), human alpha-Tubulin (mouse monoclonal, Sigma-Aldrich Cat# T6074, RRID:AB_477582) and human TSG101 (rabbit polyclonal, AbCAM, Cat # ab30871, RRID:AB_2208084) and corresponding secondary antibodies. Bound antibodies were visualized using Pierce™ ECL Western Blotting Substrate (ThermoFisher Scientific, Waltham, USA) and the chemiluminescence was measured using a BioRad ChemiDoc MP imaging system (Bio-Rad Laboratories, Inc., Hercules, USA).
Technical triplicates of Trizol-purified RNA from each experimental condition were reverse transcribed into cDNA using qScript Flex cDNA kit (Cat # 95049, Quantabio, Beverly, USA) primed with equal molar ratio of oligo-dT and random primers according to the manufacturer’s instructions. Quantitative RT-PCR was carried out using SYBR Green MasterMix (Life Technologies, Carlsbad, USA) and gene-specific primers previously validated in the literature (Table 1). These included protein-coding mRNAs EpCAM21, BIRC522, YBX123, GAPDH, and HPRT1, and lncRNAs ZFAS117, HOTAIR19, and AGAP2-AS120. Three independent experiments were performed with duplicate PCR reactions per sample. RT-qPCR data were presented as cycle threshold (CT) values. Expression values were normalized relative to GAPDH mRNA expression. Statistical analysis was performed using multiple T-test.
For this meta-analysis, the “RSEM expected count (DESeq2 standardized)” dataset was downloaded on 31st March 2020 from the TCGA_GTEx_TARGET cohort included in the UCSC Xena portal (https://xenabrowser.net/datapages/) and was manually annotated. This procedure has resulted in a dataset called “Figure 2B and C_meta_analysis_rawdata.xlsx” deposited in the DRYAD Digital Repository and used for all subsequent analyses. All data manipulations, plotting and statistical analyses were carried out in R computing environment (v 3.5.3) running in R Studio (v 1.1.414) on a Windows 10 x64 machine. The ggplot2 package (v 3.3.0) was used to render Figures 2B and 2C. Magnitude of the gene expression difference between non-tumour breast tissues and breast tumours (Hedges g effect size) was calculated using the cohen.d function included in the effsize R package (v 0.8.0). The R script containing the code for all the above computations and visualisations is available in the DRYAD Digital Repository.
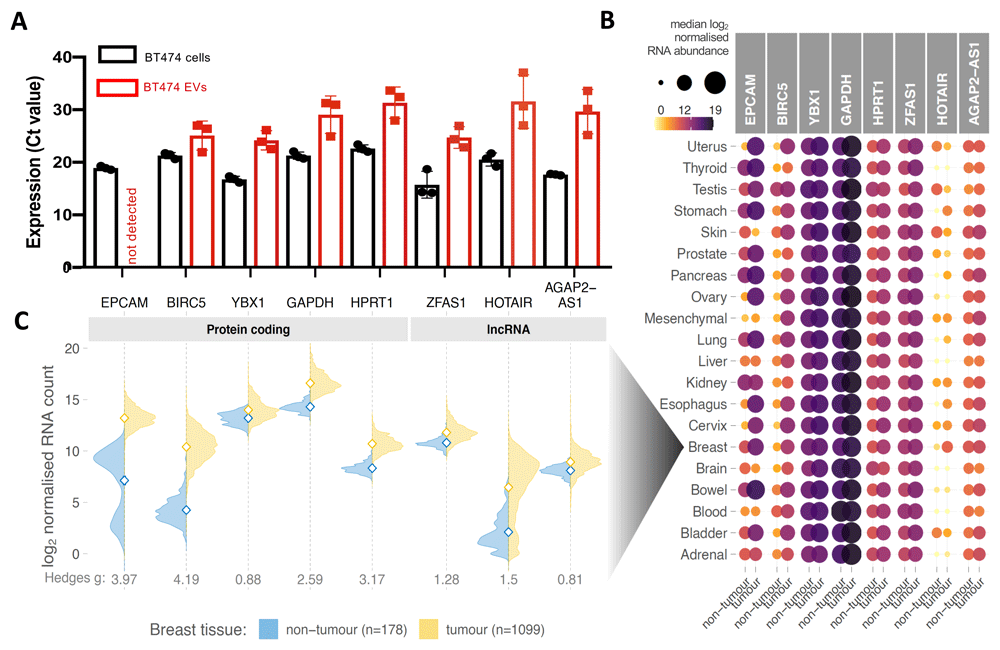
(A) Mean mRNA abundance (Ct value) of five protein-coding genes (EpCAM, BIRC5, YBX1, GAPDH, HPRT1) and three long non-coding RNAs (ZFAS1, HOTAIR, AGAP2-AS1) in BT-474 cells and their EVs. Each data point represents an average Ct value obtained in a PCR experiment using technical duplicates of an independently prepared sample. Three independent experiments were performed. Error bars denote standard errors of the mean. (B) Comparison of RNA expression of the gene panel studied in (A) between human tumours and their respective non-tumour tissues deposited in TCGA and GTEx portals. Data were manually classified into 20 different organ categories (y-axis) including 8,867 samples across 28 different cancer types and 6,874 samples across 24 non-tumour tissue types. Colour and area of the circles represent median RNA abundance; darker and larger circles indicate higher RNA expression. (C) Distribution of RNA expression of studied genes in breast tumours and breast non-cancer tissues. Open diamonds denote means of each population. Hedges g effect sizes indicate a number of standard deviations that separates the tumour and non-tumour groups. Hedges g > 0.8 demonstrates large effect size, i.e., difference between the means clearly stands out from the “noise” within the groups.
An earlier version of this article can be found on bioRxiv (doi: https://doi.org/10.1101/2020.09.27.309252).
The CELLine AD 1000 bioreactor increased the cell density and EV production due to the unique growth surfaces and fluid interactions25,27. In addition, the common issue of contaminating bovine EVs28,29 was avoided by using the serum replacement CDM-HD, which is chemically defined, protein free, and animal component free. From three independent experiments, we obtained an average of 1.9 ± 0.3 × 1011 large EVs of a mean diameter 150 ± 3 nm and 8.5 ± 0.7 × 1011 small EVs of a mean diameter 127 ± 5 nm. Negative-stained transmission electron microscope imaging showed the expected round EV morphology, and NTA size distributions resemble those seen from EVs produced in conventional culture flasks (Figure 1B–D). Low levels of contaminating proteins were observed in fractions 11–24 due to 2% CDM-HD serum replacement instead of the standard 5–10% FCS (Figure 1E). This allowed the accurate quantification of EV-associated protein markers without the concern of contaminating cellular proteins and demonstrated that the small EVs obtained using ultracentrifugation are suitable for RNA analysis.
Both the BT-474 cell lysates and BT-474 EVs of all sizes and purities isolated contained TSG101, EpCAM, and HER2 proteins (Figure 1F). Consistent with the literature, the triple-negative MDA-MB-231 breast cancer cell line did not express detectable levels of HER2 and EpCAM30. TSG101 is a regulator of the endosomal sorting and trafficking process and is expected to be present in both cells and EVs31. EpCAM is a cell adhesion glycoprotein that has been used extensively as a liquid biopsy marker for several epithelial cancers32, whilst HER2 plays an important role in breast cancer subtyping. Interestingly, HER2-positive EVs appear to increase tumour proliferation and resistance to trastuzumab therapy33.
Quantification of the abundance of several EV-associated RNAs, including protein-coding mRNAs EpCAM, BIRC5, YBX1, GAPDH, and HPRT, as well as lncRNAs ZFAS1, HOTAIR, and AGAP2-AS1, was then performed using RT-qPCR from small EVs purified by ultracentrifugation. Despite well-documented differential expression in breast cancer, EpCAM mRNA was not found to be associated with the BT-474 EVs, while BT-474 small EVs were clearly associated with established breast cancer-specific RNAs, including mRNA BIRC5 and lncRNA HOTAIR (Figure 2A). Apart from EpCAM, no significant difference (unpaired T-test) was found between cells and EVs in the RNA analysed (Figure S1).
We then explored the expression of the identical set of RNAs in 15,741 tumour and non-tumour tissue samples included in The Cancer Genome Atlas (TCGA) and Genotype Tissue Expression (GTEx) databases, respectively. Tumour and non-tumour tissues in all 20 tissues analysed expressed similar levels of YBX1, GAPDH, HPRT1, ZFAS1, and AGAP2-AS1 RNAs. The result indicates a limited use of these RNAs for differentiating tumour and non-tumour EVs. This result is consistent with the canonical “housekeeping” role of HPRT1 and GAPDH and suggests potential use of ZFAS1 and AGAP2-AS1 as housekeeping genes for analyses of lncRNAs in samples including tumour and non-tumour tissues, as well as cultured cells. Of the six candidate biomarkers investigated in this study, only BIRC522, EpCAM21 and lncRNA HOTAIR19 were found to be differentially expressed in a wide range of cancer types including breast cancer (Figure 2B and 2C).
While EVs hold promise as liquid biopsy targets for breast cancer, efficient production of EVs for molecular characterisation of EV-associated RNA can be challenging using conventional culture systems. In this technical feasibility study, we circumvented this obstacle by culturing BT-474 cells, a commonly used HER2-positive cell line, in a CELLine AD 1000 two-chambered bioreactor, which increased the cell density and EV production due to the unique growth surface and fluid interactions27. In addition, the common issue of contaminating bovine EVs29 was avoided by using the serum replacement CDM-HD, which is chemically defined, protein free, and animal component free. This bioreactor system provided highly enriched EVs in 15 mL of conditioned media, avoiding the sample loss and extra time associated with pre-centrifugation concentrators. Bioreactors were shown to improve the EV yield by over ten-fold (per volume) compared to conventional cell culture25,27,34. Cell lines including those from prostate cancer, mesothelioma, oral squamous cell carcinoma, melanoma and breast cancers were shown to grow in CELLine bioreactor25,27,34,35. Although it has been reported that cell morphology and surface markers are comparable, cells cultured in the bioreactor and conventional flasks appear to produce EVs with different metabolite content35. This could be due to 3D arrangement of cells in the bioreactor compared to monolayers in conventional flasks. The main drawback is the inability to visually observe the cells. Although the CELLine flask can be used for over 3 months of continuous cell culture, the initial cost of the CELLine flask is significantly higher than the conventional flask.
We verified that the EVs contained HER2, EpCAM, and TSG101 proteins. Transmission electron microscope imaging also allowed us to be confident that we had truly isolated small and large EVs in accordance with the MISEV guidelines36. We then demonstrated that the BT-474 small EVs were associated with lncRNAs ZFAS1, HOTAIR, and AGAP2-AS, as well as mRNAs BIRC5, YBX1, HPRT, and GAPDH using qRT-PCR.
Interestingly, the cancer-specific EpCAM mRNA was not detected in the small EVs although the EpCAM protein was detectable in the corresponding cell lysates, large EVs, and small EVs. Differential RNA expression in cancer, especially upregulation, has potential to infer a gene’s utility as a biomarker. Our finding indicates that RNAs BIRC5 and HOTAIR are promising EV-biomarkers, particularly in breast cancer, where they are substantially upregulated compared to non-tumour breast tissue. Of interest, EV associated lncRNA HOTAIR was reported to correlate with HER2-positive breast cancer37. Upregulation of serum exosomal HOTAIR was also reported to associate with poor response to chemotherapy in breast cancer patients38.
Currently, proteins dominate the EV biomarker field. However, novel EV-associated breast cancer biomarkers like lncRNAs and other RNAs are being explored more thoroughly to aid in detection and management. RNA biomarkers have higher sensitivity and specificity than proteins because PCR can amplify traces of RNA sequences with high specificity and sensitivity39. Further, it is more economical to detect RNA than protein biomarkers because each protein biomarker requires a specific antibody. These findings demonstrate the efficient production of enriched BT-474 EVs and highlight their unique cargo, especially BIRC5 mRNA and HOTAIR lncRNA. Further studies to determine their clinical significance are warranted.
DRYAD: Towards establishing extracellular vesicle-associated RNAs as biomarkers for HER2+ breast cancer. https://doi.org/10.5061/dryad.jdfn2z39340.
This project contains the following underlying data:
- Figure 1B_image_57.tif (Raw data for TEM image)
- Figure_1C_NTA_Capture_MEV_ExperimentReport.pdf (Raw data from hydrodynamic diameter distribution profiles of isolated large and small EVs measured by nanoparticle tracking analysis (NTA) with red vertical lines and blue numbers denote standard deviation and diameters at specific peaks, respectively)
- Figure_1D_NTA_Capture_SEV_ExperimentReport.pdf (Raw data from hydrodynamic diameter distribution profiles of isolated large and small EVs measured by nanoparticle tracking analysis (NTA) with red vertical lines and blue numbers denote standard deviation and diameters at specific peaks, respectively)
- Figure_1E_qEV_BCA_and_particle_data.xlsx (EV concentration determined by NTA, and protein levels determined by BCA assay of fractions acquired during separation on a qEV Original size exclusion chromatography (SEC) column)
- Figure_1F_raw_not_cropped.pptx (Raw western blot images)
- Figure 2A_RT_qPCR raw data.xlsx
For Figure 2A (Sheet 1: Raw data for RT-qPCR to examine the mRNA expression level of five protein-coding genes (EpCAM, BIRC5, YBX1, GAPDH, HPRT1) and three long non-coding RNAs (ZFAS1, HOTAIR, AGAP2-AS1) in BT-474 cells and their EVs.)
For Figure S1 (Sheet 2: Expression of RNA normalised to GAPDH to examine the mRNA expression level of five protein-coding genes (EpCAM, BIRC5, YBX1, HPRT1) and three long non-coding RNAs (ZFAS1, HOTAIR, AGAP2-AS1) in BT-474 cells and their EVs.)
- Figure 2B and C_meta_analysis_rawdata.xlsx (DeSeq2 normalised log2 (x+1) expression values of 10 genes in 8,867 tumours and 6,874 normal tissues downloaded on 31st March 2020 from the UCSC Xena portal)
The R script containing the code for all the above computations and visualisations
Data are available under the terms of the Creative Commons Zero "No rights reserved" data waiver (CC0 1.0 Public domain dedication).
The authors thank Dist. Prof. Bruce Baguley, Drs. Graeme Finlay, Marjan Askarian-Amiri, Herah Hansji and Annette Lasham for helpful discussions.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Competing Interests: No competing interests were disclosed.
Competing Interests: Co-author on a publication with Dr.Leung >10 years ago.
Reviewer Expertise: Breast biobanking, Mammary gland development, Breast cancer prevention
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Gonzalez E, Piva M, Rodriguez-Suarez E, Gil D, et al.: Human mammospheres secrete hormone-regulated active extracellular vesicles.PLoS One. 2014; 9 (1): e83955 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Breast cancer biology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Cancer and chromatin biology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: I co-authored with Dr.Leung in a publication in 2010.
Reviewer Expertise: Breast biobanking, Mammary gland development, Breast cancer prevention
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 3 (revision) 04 May 21 |
read | ||
|
Version 2 (revision) 16 Feb 21 |
read | read | |
|
Version 1 23 Nov 20 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Reviewer 1 - Nagarajan Kannan:
Extracellular vesicles (EV) are fast emerging as both therapeutic agents and biomarkers. Low ... Continue reading We thank the reviewers for their valuable and constructive feedback.
Reviewer 1 - Nagarajan Kannan:
Extracellular vesicles (EV) are fast emerging as both therapeutic agents and biomarkers. Low yields of EVs in commonly used experimental models have somewhat diminished interest and their further scrutiny. Methodologies to improve EV yield in short term cultures are desirable and much needed for this field.
In this brief communication, the authors report a culture method to enrich EV from low EV yielding breast cancer cell line, and using public datasets of tissues to bioinformatically demonstrate that these EVs are associated with RNA species enriched in tumors compared to their respective normal tissues.
The EV enrichment method appears to be simple and straightforward, and therefore the applications in cancer field is apparent. I have the following comments for the authors.
- Please provide the rate of EV production using standard 2D culture method vs your novel bioreactor-based method. It may perhaps appeal to labs working in this area to know the rate of production of EV per ml media per 1million seeded cells per hour in culture (both standard vs bioreactor).
We agree such comparison would be highly valuable but it is not trivial to accurately quantify. It is difficult to normalize the EV production per amount of cells as the cells are cultured continuously in the CELLline flasks for months with two harvests (15ml of media from the cell chamber) per week. We cannot easily remove and count the cells while they are embedded in the growth surface of the CELLline flask. In the absence of the above data, we have added to the discussion a statement that “Bioreactors were shown to improve the EV yield by over ten-fold (per volume) compared to conventional cell culture in various cell lines 6,14,21.”- Figures: It is not clear how many times these experiments were repeated or number of samples used. Please state them in the Figure Legends. Figure 1C-D showing hydrodynamic diameter distribution profiles should have intervals in Y-axis.
We apologize for the missing information. We have now added the sample size where appropriate and a number of independent experimental repeats in the figure legends. We have added ticks to the Y-axis of the hydrodynamic diameter distribution profiles in Figure 1C-D. For Figure 2A, we have now included a statement “Each data point represents an average Ct value obtained in a PCR experiment using technical duplicates of an independently prepared sample. Three independent experiments were performed. Error bars denote standard errors of the mean.”- The authors should discuss possible drawbacks to this method or choice of cell lines used in this study.
We have added the possible drawbacks and choice of cell lines used in the discussion. “Cell lines including those from prostate cancer, mesothelioma, oral squamous cell carcinoma, melanoma, and breast cancers were shown to grow in CELLine bioreactor. Although it has been reported that cell morphology and surface markers are comparable, cells cultured in the bioreactor and conventional flasks appear to produce EVs with different metabolite content. This could be due to the 3D arrangement of cells in the bioreactor compared to monolayers in conventional flasks. The main disadvantage is the inability to visually observe the cells in the CELLline bioreactor flask.- Could you further elaborate on how the TCGA data was analyzed in your methods in order to help reproduce your findings by others.
Thank you for the suggestion. We have added more details in methods describing the meta-analysis and the R script containing the code for all the above computations and visualizations is now available in the DRYAD Digital Repository.Reviewer 2 - Tracy K. Hale:
The identification of extracellular vesicle (EV) biomakers is certainly of interest in breast cancer research. This paper presents methodology to grow and isolate sufficient EVs to enable the investigation of their cargo. This study describes the growth of the breast cancer BT474 cell line in a CELLine AD 1000 bioreactor flask with media containing CDM-HD serum. This allowed the cells to be grown in 3D conditions and large numbers of EVs to be isolated without contamination from bovine EVs.
1. Figure 1 could be improved by enlarging region of Fig 1A focusing on the part of the chamber growing the cells and the isolation of EVs.
That is an excellent suggestion. We have now added an additional illustration to Figure 1A, that details a portion of the chamber flask where the cells are growing.
2. Could an antibody directed against a histone or tubulin be used in the western blot (Fig 1F) to show that the isolated EVs are not contaminated with any cellular debris?
A great idea. We have now included tubulin in Fig 1F, demonstrating that EVs were not contaminated by cellular debris.
3. In Figure 2A are the differences in HOTAIR (and the others) significant between BT474 cells and EVs? To what p-value?
We have now expressed the RNA expression values in Figure 2A as fold changes relative to GAPDH to enable comparing RNA expression in cells and EVs (Figure S1). We assume that GAPDH expression is comparable between EVs and cells. Unpaired T-tests were performed to assess the statistical significance of differential RNA expression between EVs and cells. Except for EpCAM, no significant difference in RNA expressions was found between cells and EVs.
4. The growth of cells in this system shows that there are differential levels of HOTAIR in the cells vs EVs, and gene expression data is presented to demonstrate the over expression of HOTAIR in tumours. It will be of interest to test the presence of HOTAIR in malignant vs non-malignant EVs, to support the HOTAIR as a breast cancer EV biomarker.
This is a very relevant hypothesis but testing it is beyond the scope of the current study. We have included in the discussion a statement that “Upregulation of serum exosomal HOTAIR was also reported to associate with poor response to chemotherapy in breast cancer patients (PMID: 31180050)”.
Reviewer 3 - Bruno M. Simoes:
In Figure 1 – Experiments comparing the results with conventional 2D culture system should be shown to support the statements in the paper. And comparison using bovine serum should be shown too.
This is a great point that has also been raised by Referee 1, and we have discussed the caveats of accurately comparing the EV production in the bioreactor and conventional flasks in our response to Referee 1. As for the comparison between CDM-HD serum replacement and bovine serum, this comparison is not included in the current study as bovine EVs from regular FCS has previously been shown to contaminate EV preparations from mammalian cells (DOI: 10.1039/C7NR08360B).
Use of specific EV markers is needed to confirm the presence of EVs – see Gonzalez et al, Plos One, 20141.
We have now included tubulin in Fig 1F, demonstrating that EVs were not contaminated by cellular debris. We are convinced that excluding the presence of cellular debris in the EV preparation is essentially equivalent to confirming the presence of EVs by specific markers.
The use of at least another cell line is warranted to validate the data and the use of primary cells would be ideal to assess potential clinical translation of the data.
Yes, we agree. We are in the process of characterising the EV protein expression profile in several other cell lines cultured in Bioreactor flasks for another project. These data will be published as a separate manuscript.
Figure 2A – The rational for selection of this list of EV-associated RNAs is not clear and needs to be defined. Real-time PCR data should be normalized to a reference transcript.
Also, it is not clear if these experiments were done with large EVs, small EVs or both? If data corresponds only to small EVs then the experiments should be repeated with large EVs too.
Thanks for these great suggestions for improvement. The RNAs studied in our paper were selected on the basis of their association with HER2+ breast cancer. We have now normalized the RNA expression with GAPDH and expressed the data as fold changes relative to GAPDH (Figure S1). The qPCR experiments in Figure 2A were conducted with small EVs purified by ultracentrifugation (Figure 1D) as we have now indicated in the manuscript. As the small EVs purified by ultracentrifugation also contains a small number of large EVs, the RNA expression data could not be reliably associated with large EVs. Therefore, we argue that conducting qPCR experiments on large EVs may generate confusing and hard to interpret results. Large EVs are difficult to purify due to their large size and can contain plenty of cellular debris when purified by ultracentrifugation.
Authors state “RNAs BIRC5 and HOTAIR are promising EV-biomarkers” but this is not proven by the data. The housekeeping genes GAPDH and HPRT are also expressed in EVs. Testing the presence of BIRC5 and HOTAIR RNAs in tumour vs non-tumour EVs is warranted.
We base this statement on the bioinformatics analysis presented in Figure 2C. BIRC5 and HOTAIR satisfy two requirements for a suitable biomarker. i) They are expressed higher in breast cancer relative to non-tumour breast tissue. ii) Normal tissues express the lowest levels of both genes (especially HOTAIR) compared to other RNAs studied. This low RNA abundance in normal tissues presents a potential benefit in differentiating their cancer-associated RNA expression.
Testing the presence of BIRC5 and HOTAIR RNAs in tumour vs non-tumour from plasma is a superb suggestion and is being pursued in collaboration with another research group.
Number of independent experiments and replicates needs to be stated throughout the manuscript.
We have included in Figure 1 legend "Representative images/data from three independent experiments were shown in B– F." and Figure 2A legend “Each data point represents an average Ct value obtained in a PCR experiment using technical duplicates of an independently prepared sample. Three independent experiments were performed. Error bars denote standard errors of the mean.”
Reviewer 1 - Nagarajan Kannan:
Extracellular vesicles (EV) are fast emerging as both therapeutic agents and biomarkers. Low yields of EVs in commonly used experimental models have somewhat diminished interest and their further scrutiny. Methodologies to improve EV yield in short term cultures are desirable and much needed for this field.
In this brief communication, the authors report a culture method to enrich EV from low EV yielding breast cancer cell line, and using public datasets of tissues to bioinformatically demonstrate that these EVs are associated with RNA species enriched in tumors compared to their respective normal tissues.
The EV enrichment method appears to be simple and straightforward, and therefore the applications in cancer field is apparent. I have the following comments for the authors.
- Please provide the rate of EV production using standard 2D culture method vs your novel bioreactor-based method. It may perhaps appeal to labs working in this area to know the rate of production of EV per ml media per 1million seeded cells per hour in culture (both standard vs bioreactor).
We agree such comparison would be highly valuable but it is not trivial to accurately quantify. It is difficult to normalize the EV production per amount of cells as the cells are cultured continuously in the CELLline flasks for months with two harvests (15ml of media from the cell chamber) per week. We cannot easily remove and count the cells while they are embedded in the growth surface of the CELLline flask. In the absence of the above data, we have added to the discussion a statement that “Bioreactors were shown to improve the EV yield by over ten-fold (per volume) compared to conventional cell culture in various cell lines 6,14,21.”- Figures: It is not clear how many times these experiments were repeated or number of samples used. Please state them in the Figure Legends. Figure 1C-D showing hydrodynamic diameter distribution profiles should have intervals in Y-axis.
We apologize for the missing information. We have now added the sample size where appropriate and a number of independent experimental repeats in the figure legends. We have added ticks to the Y-axis of the hydrodynamic diameter distribution profiles in Figure 1C-D. For Figure 2A, we have now included a statement “Each data point represents an average Ct value obtained in a PCR experiment using technical duplicates of an independently prepared sample. Three independent experiments were performed. Error bars denote standard errors of the mean.”- The authors should discuss possible drawbacks to this method or choice of cell lines used in this study.
We have added the possible drawbacks and choice of cell lines used in the discussion. “Cell lines including those from prostate cancer, mesothelioma, oral squamous cell carcinoma, melanoma, and breast cancers were shown to grow in CELLine bioreactor. Although it has been reported that cell morphology and surface markers are comparable, cells cultured in the bioreactor and conventional flasks appear to produce EVs with different metabolite content. This could be due to the 3D arrangement of cells in the bioreactor compared to monolayers in conventional flasks. The main disadvantage is the inability to visually observe the cells in the CELLline bioreactor flask.- Could you further elaborate on how the TCGA data was analyzed in your methods in order to help reproduce your findings by others.
Thank you for the suggestion. We have added more details in methods describing the meta-analysis and the R script containing the code for all the above computations and visualizations is now available in the DRYAD Digital Repository.Reviewer 2 - Tracy K. Hale:
The identification of extracellular vesicle (EV) biomakers is certainly of interest in breast cancer research. This paper presents methodology to grow and isolate sufficient EVs to enable the investigation of their cargo. This study describes the growth of the breast cancer BT474 cell line in a CELLine AD 1000 bioreactor flask with media containing CDM-HD serum. This allowed the cells to be grown in 3D conditions and large numbers of EVs to be isolated without contamination from bovine EVs.
1. Figure 1 could be improved by enlarging region of Fig 1A focusing on the part of the chamber growing the cells and the isolation of EVs.
That is an excellent suggestion. We have now added an additional illustration to Figure 1A, that details a portion of the chamber flask where the cells are growing.
2. Could an antibody directed against a histone or tubulin be used in the western blot (Fig 1F) to show that the isolated EVs are not contaminated with any cellular debris?
A great idea. We have now included tubulin in Fig 1F, demonstrating that EVs were not contaminated by cellular debris.
3. In Figure 2A are the differences in HOTAIR (and the others) significant between BT474 cells and EVs? To what p-value?
We have now expressed the RNA expression values in Figure 2A as fold changes relative to GAPDH to enable comparing RNA expression in cells and EVs (Figure S1). We assume that GAPDH expression is comparable between EVs and cells. Unpaired T-tests were performed to assess the statistical significance of differential RNA expression between EVs and cells. Except for EpCAM, no significant difference in RNA expressions was found between cells and EVs.
4. The growth of cells in this system shows that there are differential levels of HOTAIR in the cells vs EVs, and gene expression data is presented to demonstrate the over expression of HOTAIR in tumours. It will be of interest to test the presence of HOTAIR in malignant vs non-malignant EVs, to support the HOTAIR as a breast cancer EV biomarker.
This is a very relevant hypothesis but testing it is beyond the scope of the current study. We have included in the discussion a statement that “Upregulation of serum exosomal HOTAIR was also reported to associate with poor response to chemotherapy in breast cancer patients (PMID: 31180050)”.
Reviewer 3 - Bruno M. Simoes:
In Figure 1 – Experiments comparing the results with conventional 2D culture system should be shown to support the statements in the paper. And comparison using bovine serum should be shown too.
This is a great point that has also been raised by Referee 1, and we have discussed the caveats of accurately comparing the EV production in the bioreactor and conventional flasks in our response to Referee 1. As for the comparison between CDM-HD serum replacement and bovine serum, this comparison is not included in the current study as bovine EVs from regular FCS has previously been shown to contaminate EV preparations from mammalian cells (DOI: 10.1039/C7NR08360B).
Use of specific EV markers is needed to confirm the presence of EVs – see Gonzalez et al, Plos One, 20141.
We have now included tubulin in Fig 1F, demonstrating that EVs were not contaminated by cellular debris. We are convinced that excluding the presence of cellular debris in the EV preparation is essentially equivalent to confirming the presence of EVs by specific markers.
The use of at least another cell line is warranted to validate the data and the use of primary cells would be ideal to assess potential clinical translation of the data.
Yes, we agree. We are in the process of characterising the EV protein expression profile in several other cell lines cultured in Bioreactor flasks for another project. These data will be published as a separate manuscript.
Figure 2A – The rational for selection of this list of EV-associated RNAs is not clear and needs to be defined. Real-time PCR data should be normalized to a reference transcript.
Also, it is not clear if these experiments were done with large EVs, small EVs or both? If data corresponds only to small EVs then the experiments should be repeated with large EVs too.
Thanks for these great suggestions for improvement. The RNAs studied in our paper were selected on the basis of their association with HER2+ breast cancer. We have now normalized the RNA expression with GAPDH and expressed the data as fold changes relative to GAPDH (Figure S1). The qPCR experiments in Figure 2A were conducted with small EVs purified by ultracentrifugation (Figure 1D) as we have now indicated in the manuscript. As the small EVs purified by ultracentrifugation also contains a small number of large EVs, the RNA expression data could not be reliably associated with large EVs. Therefore, we argue that conducting qPCR experiments on large EVs may generate confusing and hard to interpret results. Large EVs are difficult to purify due to their large size and can contain plenty of cellular debris when purified by ultracentrifugation.
Authors state “RNAs BIRC5 and HOTAIR are promising EV-biomarkers” but this is not proven by the data. The housekeeping genes GAPDH and HPRT are also expressed in EVs. Testing the presence of BIRC5 and HOTAIR RNAs in tumour vs non-tumour EVs is warranted.
We base this statement on the bioinformatics analysis presented in Figure 2C. BIRC5 and HOTAIR satisfy two requirements for a suitable biomarker. i) They are expressed higher in breast cancer relative to non-tumour breast tissue. ii) Normal tissues express the lowest levels of both genes (especially HOTAIR) compared to other RNAs studied. This low RNA abundance in normal tissues presents a potential benefit in differentiating their cancer-associated RNA expression.
Testing the presence of BIRC5 and HOTAIR RNAs in tumour vs non-tumour from plasma is a superb suggestion and is being pursued in collaboration with another research group.
Number of independent experiments and replicates needs to be stated throughout the manuscript.
We have included in Figure 1 legend "Representative images/data from three independent experiments were shown in B– F." and Figure 2A legend “Each data point represents an average Ct value obtained in a PCR experiment using technical duplicates of an independently prepared sample. Three independent experiments were performed. Error bars denote standard errors of the mean.”