Keywords
miRNA, glia, Drosophila, screens, GBF1
This article is included in the NC3Rs gateway.
miRNA, glia, Drosophila, screens, GBF1
We have now modified some aspects of our text to explain better some points highlighted by the reviewers. This includes revisions to Figures 1 and 3 to incorporate one of the reviewers comments on the visibility of labels and of the items identified by arrows.
See the authors' detailed response to the review by Jeff W. Barclay
See the authors' detailed response to the review by Ivana Bjedov
This screen has provided a thorough analysis of glial functions in ageing
Potential to shortcut gene discovery through miRNAs effect. The screening of only ~200 mutant lines potentially targets >6000 genes.
Potential to identify complex regulatory networks that include miRNAs and target genes.
Validated the identification of essential genes for the adult nervous system and their functions specifically in motor control.
An open-access searchable database for future discoveries upon improved precision of miRNA-target predictions.
The searchable database can be easily updated upon emergence of updated miRNA target predictions.
RNAi lines are publicly available from the Vienna Drosophila Resource Centre stock collection.
Genetic studies in Drosophila are quicker and more sophisticated compared to vertebrate studies. They also maintain high conservation of functions.
Despite the fact that glial cells were initially identified simply as the connective tissue of the brain1, work developed in the past decades has shed a light on a much more intricate role for these cells in developing and maintaining nervous system homeostasis (reviewed in 2). From neuronal nutrient supply3, to neurotransmitter recycling4–6, to being the first line of immune response in the brain7, glial cells have been shown to actively contribute to the correct functioning of the brain.
More recently, several studies have been taking advantage of Drosophila’s powerful genetic manipulation to better understand the role of glia in the development and maintenance of the nervous system (see 8 for review).
The use of invertebrate models is also a powerful 3Rs solution to reduce and replace animal experiments. It expressly applies to complex matters in which cross-talk between different cell types (e.g. glia and neurons) is a focal point of the investigation, given that these complex environments are more difficult to model in vitro and in silico. Popular animal models for studying glial functions are zebrafish, which provide a useful platform for tissue and cell biology, with some capability for genetic manipulation9 and genetically modified mice10. Despite having a different developmental origin, glial cells have converged in Drosophila and mammals towards the same key functions of neurotransmission regulation, insulation and immune surveillance/phagocytosis8, making the fruit-fly an organism of choice for studying the function of glial cells.
We have tackled the functions of glial cells in ageing. We have previously screened a large collection of miRNAs regarding their effects on Drosophila’s lifespan upon ectopic expression in glial cells in adult flies and have validated this screen through the analysis of repo, an already-established key glia gene11. The experimental advantage of performing a miRNA-based screen followed by in silico identification and ranking of predicted miRNAs target transcripts11,12 has, however, its bottleneck in the validation of the action of the genes of interest. In principle, the specific knockdown of predicted target genes should mimic, to some extent, the phenotype obtained upon corresponding miRNA overexpression.
In fact, using databases of predicted miRNA-target genes previously allowed us to identify repo as an important player for maintaining glial function and, consequently, homeostasis in the adult brain11. We have shown that while the miR-1-repo axis is physiologically relevant only in the embryo during the glia versus haemocyte cell fate choice13, the miRNA-target relationship can be exploited as a discovery tool to identify the functions of a target gene in a different context, namely adult glial functions11.
While the focus on repo was based on its already-established role in glia cell function, here we attempt a global and unbiased systematic in silico approach. In order to systematically identify potential target genes that could account for the lifespan phenotype, focusing on the miRNAs that shortened lifespan, we set out to devise a quantitative algorithm. The aim of this algorithm is to identify and rank the predicted target genes so that those ranking on top would be the most relevant for adult glia in lifespan and ageing.
This is followed by experimental validation of the function of these targets in adult glia in the same paradigm used in the miRNAs screen.
We conclude that this approach is valid but has issues of efficiency given the large number of predicted targets that do not recapitulate the expected phenotype. We also establish that there is no significant synergy generated by focusing on the common predictions between all available miRNAs target databases. Nevertheless, the main outcome of our work is a list of candidate genes whose function is essential in glial cells during ageing. These genes can be studied in the future in Drosophila, with the tools identified here, rather than in genetically modified mouse models or in zebrafish, providing an incentive towards animal replacement and reduction and advancing the 3Rs. Mouse and zebrafish neuroscientists and geneticists could take advantage of this information to test preliminary approaches and exploratory experiments in Drosophila, prior to validation in their system reducing the number of animals used. Alternatively, they may entirely replace vertebrate animals with Drosophila to study highly conserved genes and glial functions.
The success of this in silico approach is exemplified by our analysis of one of the top predicted targets: gartenzwerg (garz), the fly orthologue of GBF1 (golgi brefeldin A resistant guanine nucleotide exchange factor 1), a small GTPase guanine exchange factor. Here, we show that garz is an essential factor in glia homeostasis maintenance.
Small GTPases regulate a wide range of cellular events such as proliferation, morphology, nuclear transport and vesicle formation14. The conversion from GDP-bound (inactive) to GTP-bound (active) forms of these enzymes relies on the activity of GTPase activating proteins (GAPs) and guanine nucleotide exchange factors (GEFs). While GAPs are responsible for their inactivation through GTP hydrolysis, GEFs are responsible for their activation promoting the exchange of GDP by GTP15.
GEFs belonging to the Sec7 domain protein family are responsible for the activation of Arf (ADP-ribosylation factor) GTPases which are associated with the recruitment of coat proteins (COP) to vesicle budding sites16–18. GBF1 is part of this family19 and is highly conserved in all eukaryotes, conferring significant translatability of the findings obtained using different model organisms.
Strongly localized in the cis-Golgi compartment, GBF1 has been shown to regulate vesicle trafficking between the endoplasmic reticulum (ER) and the Golgi apparatus20–24. Mutated versions or knock-down of garz expression brings about epithelial morphogenesis defects during development conditioning embryonic trachea and larval salivary gland formation20,21. Additionally, in accordance with a role in membrane delivery and vesicular trafficking, silencing of garz in these glands impairs membrane delivery of adhesion molecules25. Independently from its role in secretion, GBF1/garz has also been implicated in pinocytosis26; intestinal stem cell survival27; cell cycle28,29; unfolded protein response events29; mitochondria morphology and function30; and autophagy31,32.
Here we show that garz knock-down resulted not only in lifespan reduction but also in motor deficits of adult flies and in subcellular phenotypes indicative of dysfunctions in trafficking, autophagy and mitochondria. Additionally, miRNAs overexpression and garz knockdown phenotypes were reverted by expression of its mammalian orthologue GBF1, stressing the conservation of functions and the appropriateness of using Drosophila in place of vertebrate models to study the biology of GBF1.
The following databases were used for the prediction of miRNA targets:
MicroCosm (https://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/)
microRNA.org (http://www.microrna.org/microrna/home.do)
TargetScan (http://www.targetscan.org/fly_72/)
PicTar (https://pictar.mdc-berlin.de/)
Each of the databases provides for every miRNA a numerical prediction of the likelihood of targeting a given gene (Score). For MicroCosm and PicTar this was used without additional steps. In the case of miRNA.org this score is a negative value and we have squared it to obtain a positive number. In the case of TargetScan a numerical score was calculated on the basis of the information provided by the database as follows: conserved 8mer = 10 points, conserved 7mer-m8 = 6 points, conserved 7mer-1A = 4 points, poorly conserved 8mer = 8 points, poorly conserved 7mer-m8 = 4 points and poorly conserved 7mer-1A = 2 points. A detailed explanation of the 8mer and 7mer species can be found on the TargetScan website and in the original publication33.
The algorithm for ranking targets within each database consists of two steps.
Step 1 - column (Score)*Av(χ2) or (Score2)*Av(χ2):
For each miRNA, every target score (or its square value) was multiplied by the Average Chi square (χ2) obtained in the miRNAs screen (from Table 1). Information regarding different mRNAs for the same gene, where available, was grouped under the same gene name
Step 2 - column Σ(Score)*Av(χ2)] or Σ(Score2)*Av(χ2)]:
For each target gene, as defined by its CG number/accession ID, all values resulting from all miRNAs predicted to target the same gene were summed in a final ranking value. Information regarding different mRNAs from the same gene, where available, was grouped under the same gene name.
The algorithm for comparing the ranking between different databases and providing a final common ranking consists of two steps:
Step1 - column Normalised Σ[(Score)*Av(χ2)] or Normalised Σ[(Score2)*Av(χ2)]
For each database the Σ(Score)*Av(χ2)] was normalised to 100 and then weighted for the fraction of miRNAs present in the database, out of the total tested in our miRNAs screen. For TargetScan the groups of miRNAs families were counted as one unit in each case.
Step 2 – column Σ{Normalised Σ[(Score(2))*Av(χ2)]} For each target gene, all values from all databases were summed into a final ranking number.
To determine the strength of miRNAs in our lifespan assay we have used the averaged χ2 values from each transgenic line used in our previously published analysis11. When only one line was tested for a given miRNA, the value was divided in half, i.e. assuming a neutral value of 0 for a second putative untested line. For the TargetScan database, some miRNAs are grouped in families requiring an amendment to our approach. In this case, we have averaged all miRNAs in the given families. Additionally, some of the lines tested for these grouped miRNAs had, in the original screen the opposite effect of what is here considered, i.e. extending lifespan with respect to the control used. To account for this opposite effect the χ2 values for these miRNAs have been given negative values and have been effectively subtracted, when calculating the Av(χ2) parameter.
Flies were kept on standard cornmeal agar food (0.8% w/v agar, 2% w/v cornmeal, 8% w/v glucose, 5% w/v Brewer’s yeast, 1.5% v/v ethanol, 0.22% v/v methyl- 4-hydroxybenzoate, 0.38% v/v propionic acid) at 18°C or room temperature. Unless stated otherwise, w1118 flies were used as control. The following lines were acquired from the Bloomington collection: w1118 (RRID:BDSC_3605), repo-Gal4 (RRID:BDSC_7415), NP2222-Gal4 (RRID:DGGR_112830), moody-Gal4, elav-Gal4 (RRID:BDSC_8765), tub-Gal80ts (RRID:BDSC_7019). alrm-Gal4 (RRID:BDSC_67031) was kindly provided by M. Freeman (University of Massachusetts) ; UAS-miR-1, UAS-miR-79 and UAS-miR-315 were generated by E. Lai (Sloan Kettering Institute) for the miR library34; UAS-garz-RNAi (42140/GD and 42141/GD) as well as all RNAi lines used are from Vienna Drosophila Resource Center (VDRC); gliotactin-Gal4 was provided by R. Sousa-Nunes; UAS-mito-GFP was provided by J. Bateman; UAS-garz; UAS-garzSec7-; UAS-GBF1 and UAS-ΔGBF1Sec7- were kindly provided by S. Luschnig.
Lifespan analysis was performed as previously described35. Briefly, crosses were maintained at 18°C throughout the whole development of the progeny. Within the first 5 days post-eclosion, adult flies were collected, and equal numbers of female and male flies were pooled together. An equal number of flies was distributed in three vials, a total of 60 flies was used. This group size has a power of 0.8 in one tailed survival test at 50% survival for the control group and 29% for an experimental group at 0.05 significance. Lifespan assessment was performed in a controlled environment of 29°C and 60% humidity, three times a week. Upon short CO2 anaesthesia (5 s), the number of dead vs alive flies was counted, and the alive flies transferred into a fresh vial.
Single fly tracking was carried out as previously described11. In each experiment, up to 20 flies per genotype were placed into individual glass tubes. This group size has a power of 0.9 and significance 0.05 for three groups with an effect size of 0.48, as measured for the mean bout length. All the genotypes were positioned on the same platform, having two shaft-less motors placed underneath each subplatform containing each, one genotype. The protocol used consisted of 6 stimuli events equally split during a period of 2 h and 15 min, the first one starting after 30 min of recording and the last one 30 min before the end of the protocol. Each stimuli event was composed of 5 vibrations of 200 ms spaced by 500 ms. The x/y position of each single fly was tracked and analysed using DART software 1.0 (freely distributed upon request to info@bfklab.com) in order to evaluate the relative speed and activity before, during and after the stimuli event. The speed analysis was used for the “Stimuli Response Trace” and the general activity used to deduce “Active Speed”, “Mean Bout Length” and “Inter-Bout Interval”, using a custom-made modification of the DART software36. Raw data were analysed with GraphPad Prism for statistical significance and DART-derived graphs were edited with Adobe Illustrator CC2017 (RRID:SCR_010279).
Flies (N=5–10) were briefly (5 s) anesthetized with CO2 and kept on ice, entire fly brains were dissected under a stereoscope and immediately fixed in 4% paraformaldehyde (PFA, from EMS) in Phosphate Buffer Saline (PBS) for 30 min. After washing with PBS, the brains were incubated for blocking in PBS with 0.3% triton-X (BDH 306324N) (PBT) and 10% foetal bovine serum (Sigma F4135) for 1 hr. Primary antibody incubation was done overnight at 4°C and followed by three washes (20 min each) in PBT. Secondary antibody incubation for 1hr at room temperature was followed by three washes. All steps were in 50-µl volume in a 96-well plate on a gentle rocker. Brains were then mounted on a slide in Vectashield with DAPI (Vector Labs). The following primary antibodies, diluted in blocking solution (see above): anti-Repo (1/100, mouse DSHB 8D12, RRID:AB_528448); anti-GFP (1/1000, rabbit, Life technologies, A11122) anti-GFP(1/100, mouse, Roche, RRID:AB_390913), anti-GFP (1/500, chicken, kindly provided by M. Meyer); anti-Ref(2)P (1/2000, rabbit, a gift of Tor Erik Rusten). Secondary antibodies were all from Life technologies (conjugated with Alexa-488, Alexa-555 or Alexa-666) and diluted 1/200 in blocking solution (see above).
Z-stacks at intervals of 0.3 µm or 5 µm were taken at 1024×1024 pixel/inch resolution. For control vs garzIR comparisons, microscope settings were established using control flies to have a GFP signal below saturation and kept unchanged throughout all acquisitions. All images were acquired with a Leica TCS SP5 confocal microscope and mitochondria sphericity, volume and surface area in Figure 3B,C were measured using the 3D Object Counter 2.0.1 plugin37 in the ImageJ Fiji 1.52n software (RRID:SCR_002285).
All statistical analysis was performed with Graph- Pad Prism 7 software (RRID:SCR_002798). For all lifespans, the statistical analysis was performed using the log–rank test of the Kaplan and Meier method. For behavioural experiments (DART), the statistical analysis was done by one-way ANOVA using Dunnett’s multiple comparisons post hoc test. Significance is shown by asterisks in all figures as follows: *P<0.05, **P<0.01, ***P<0.001, and ****P<0.0001.
Firstly, such algorithm should prioritise the information for the miRNAs that had the strongest effect on the fly lifespan in our miRNA screen. To achieve this, we have quantified the average strength of each miRNA using the Chi square (χ2) of each Kaplan Mayer analysis (Table 1).
To identify potential target genes, we used four different databases available online: EBI MicroCosm, PicTar, microRNA.org and TargetScan. Each database weights the likelihood of every miRNA to target a given gene with a numerical score. Where this is different, for TargetScan, we calculated a numerical score on the basis of the sequence information provided by the database (see Methods).
Therefore, to rank target genes within each database taking into account both the likelihood of being targeted by a given miRNA and the strength of the effect of this miRNA in adult glia, we first multiplied the average strength of each miRNA from our screen (values in Table 1) by the strength of the target prediction (Score) given by the database, obtaining the parameter (Score)*Av(χ2). This was done for all miRNAs tested in our screen that were present in each database.
Because a given gene can be targeted by more than one miRNA, to rank its overall importance in adult glia, we have summed all the values obtained for a given gene that were calculated for different miRNAs, obtaining the parameter Σ[(Score)*Av(χ2)]. In the case of TargetScan, some miRNAs are grouped in families and we have considered them as a single unit value. This underweights these miRNAs in comparison to others and the genes targeted by them (for instance a gene targeted by miR-9a, miR-9b and miR-9c would obtain a Σ[(Score)*Av(χ2)] that is the sum of three (Score)*Av(χ2) in the other databases, but for TargetScan it would only reflect one (Score)*Av(χ2). Our reasoning was that grouped miRNAs in TargetScan was not taking into account valuable information and this should be reflected in a penalisation in the ranking.
In conclusion we have ranked target genes according to Σ[(Score)*Av(χ2)] for EBI MicroCosm (Extended data Table 1)38, PicTar (Extended data Table 2)38, microRNA.org (Extended data Table 3)38 and TargetScan (Extended data Table 4)38. Surprisingly, this revealed that there was very little agreement among the four databases. The top-ranking genes obtained using the same algorithm were very different and only 5.6% (i.e. 520 genes) of target predictions were common to all four databases (Figure 1A).
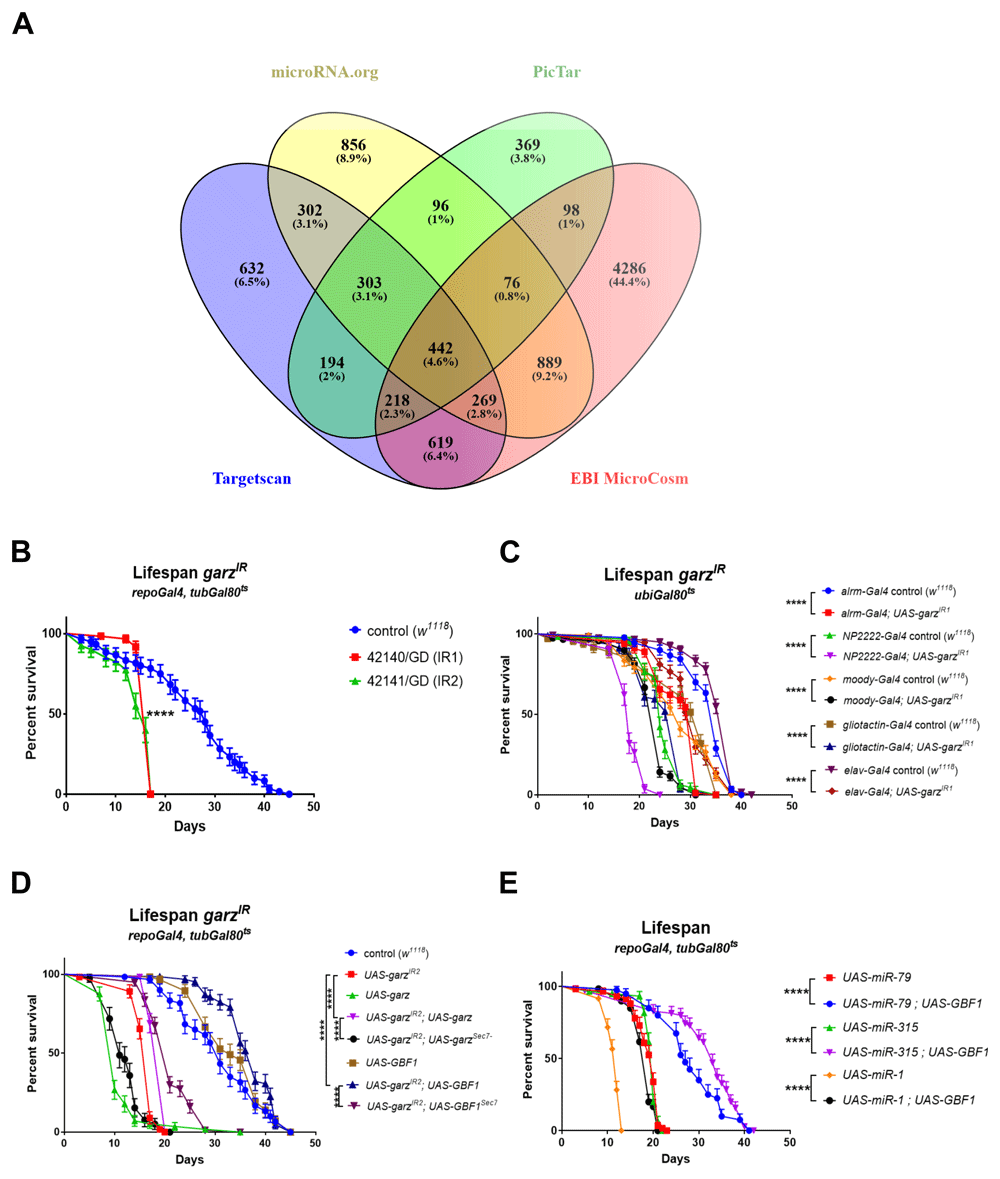
(A) Venn diagram referring to the data in Table 2 and illustrating the overlap between the four different databases used to predict gene targets of the miRNAs whose expression in the adult glia resulted in a significant reduction in fly lifespan. Only 520 target genes are in common among all four databases, garz falls in this group. A remarkably large number of genes as targets were uniquely predicted by the MicroCosm database. (B) Two RNAi lines against garz bring about a very significant reduction in fly lifespan in comparison to controls, when expressed in all adult glia. N=60 for each genotype, Error bars SEM, pairwise comparisons: Log-rank (Mantel-Cox) test. The full dataset can be accessed at DOI 10.17605/OSF.IO/8E3NS as part of Table 3. (C) Knock-down of garz in sub-populations of glial cells, astrocyte-like (alrm-Gal4), Cortex glia (NP2222-Gal4), sub-perineural glia (moody-Gal4), perineural and PNS glia (gliotactin-gal4) or in neurons (elav-Gal4) brings about a significant reduction in lifespan in comparison to controls. N=60 for each genotype, Error bars SEM, pairwise comparisons: Log-rank (Mantel-Cox) test. The full dataset can be accessed at DOI 10.17605/OSF.IO/HQCDG. (D) Lifespan reduction due to RNAi against garz in adult glia is rescued by an exogenous UAS-garz transgene and by a transgene expressing the human orthologue GBF1 under UAS control. Note that overexpression of garz in an otherwise wt background is highly detrimental to fly lifespan, whereas overexpression of GBF1 in a wt background has no adverse effects. Mutations leading to a non-functional Sec7 domain eliminate or drastically reduce the ability of garz or GBF1 transgenes to rescue fly lifespan. N=60 for each genotype, Error bars SEM, pairwise comparisons: Log-rank (Mantel-Cox) test. The full dataset can be accessed at DOI 10.17605/OSF.IO/5RGEF. (F) Co-expression of human GBF1 significantly extends the short lifespan caused by overexpression of miR-1, miR-79 and miR-315 in adult glia. N=60 for each genotype, Error bars SEM, pairwise comparisons: Log-rank (Mantel-Cox) test. The full dataset can be accessed at DOI 10.17605/OSF.IO/B37DF.
To rank these common targets for their predicted overall relevance in adult glia in ageing, we have devised additional steps. First, to make the numerical rankings from each database comparable, we have calculated the Normalised Σ[(Score)*Av(χ2)] parameter by normalising the maximum value to 100. Additionally, we have weighted this number for the fraction of miRNAs present in each database, out of the total tested in our miRNAs screen. Out of 44 miRNAs screened, 31 were present in EBI Microscosm, 28 in PicTar, 43 in microRNA.org and 40 in TargetScan. The rationale for this weighting was to prioritise the databases carrying more information that was relevant to our screen. Then, for each target gene, we have combined all these scores from the four databases generating the final parameter Σ{Normalised Σ[(Score(2))*Av(χ2)]} for all targets, including the 520 that were commonly predicted by all databases (Table 2).
The ranking scores from all four databases were pooled to obtain a global rank of all targets predicted by our analysis and a list of targets that are predicted by all four databases. Because the different databases contained information about some, but not all, miRNAs analysed in our screen we have weighted the completeness of each database by normalising the Σ[(Score)*Av(χ2)] by the fraction of miRNAs listed in the database, out of the ones tested in our screen. In addition, to make the ranking from each database equally valued in this analysis, we have normalised each score to 100 as a maximum possible value for each database – column Normalised Σ[(Score)*Av(χ2)]. Thereafter, all values for each target have been added – column Σ{Normalised Σ[(Score)*Av(χ2)]} – for each target and for a specific list of 520 targets that have been predicted by all four databases, albeit with different scores. Only the top 30 rows are shown here. The full table can be accessed at https://doi.org/10.17605/OSF.IO/QWUAY.
Strength of the effect on fly lifespan of RNAi lines against some of the gene targets predicted by all four databases. For most genes two different RNAi lines have been tested. In red are the lines that, in agreement with the miRNA prediction, shorten the lifespan, in comparison to controls (w1118) when specifically expressed in the adult flies with repo-Gal4 and tub-Gal80ts. In green are the lines that have the opposite effect and prolong lifespan. In black are the lines that had no effect. Highlighted in yellow are the genes for which all lines tested had the same effect and shortened lifespan. Highlighted in pink is the gene for which all lines tested had the same effect and prolonged the lifespan. To combine the target RNAi strength with the strength of the prediction we have averaged the χ² for each miRNA according to the same rules followed in Table 1 and multiplied it for the final score from Table 2 – column Σ{Normalised Σ[(Score)*Av(χ²)] *{Av(χ²)IR}. The top rank was achieved by garz, which was also one of the 6 genes with all RNAi lines tested having the same effect. We have also repeated the same procedure separately for the four different databases using the final normalised score (Normalised Σ[(Score)*Av(χ²)]) obtained from each database in Tables 2, 3, 4 and 5, and also reported in Table 2– columns Normalised Σ[(Score)*Av(χ²)] *{Av(χ²)IR} for each database. The predicting power for the combination of all databases and for each database sums all scores in this column to quantify the global predicting value of each database in comparison to their combination. This is further normalised by the number of predicted targets for the whole screen, to measure the efficiency of each database when predicting target genes. The full dataset can be accessed at DOI 10.17605/OSF.IO/8E3NS.
Strength of the effect on fly lifespan of RNAi lines against some of the gene targets predicted by some, but not all, databases. In red are the lines that, in agreement with the miRNA prediction, shorten the lifespan, in comparison to controls (w1118) when specifically expressed in the adult flies with repo-Gal4 and tub-Gal80ts. In green are the lines that have the opposite effect and prolong lifespan. In black are the lines that had no effect. Highlighted in yellow is the gene for which all lines tested had the same effect and shortened lifespan. To combine the target RNAi strength with the strength of the prediction we have averaged the χ² for each miRNA according to the same rules followed in Table 1 and multiplied it for the final normalised score (Normalised Σ[(Score)*Av(χ²)]) obtained from each database in Extended data Tables 1, 2, 3and 4 [49], and also reported in Table 2 – columns Normalised Σ[(Score)*Av(χ²)] *{Av(χ²)IR} for each database. The predicting power for each database sums all scores in this column to quantify the global predicting value of each database. This is further normalised by the number of predicted targets for the whole screen, to measure the efficiency of each database when predicting target genes, as in Table 3. The full dataset can be accessed at DOI 10.17605/OSF.IO/QTASN.
To test these predictions, we decided to screen for the lifespan effect, a number of RNAi lines from Vienna Drosophila Resource Center (VDRC) that were already present in our stock collection. These corresponded to a random selection of approximately 10% (51 out of 520) of commonly predicted target genes. Adopting a similar strategy used for the miRNA screen, we have used the repo-Gal4, tub-Gal80ts inducible system to trigger the RNAi expression in all glial cells in adult flies. As negative control, we used the offspring of crossing repo-Gal4, tub-Gal80ts to w1118 throughout the screen. The expectation was that RNAi against these target genes in adult glia, would phenocopy the effect of the miRNAs that are predicted to target them, therefore shortening lifespan.
The gold standard commonly used by the Drosophila community to gain confidence about the effects of RNAi knock-down is to obtain a similar effect when testing two RNAi lines against the same gene (2-RNAi lines criterion). Remarkably, only in six cases at least two different RNAi lines tested for the same gene delivered the shorter lifespan phenotype that was predicted (Table 3). In another case both RNAi lines tested had the same effect, but it was the opposite of the predicted one, extending lifespan with respect to the control flies.
In other cases (11/51) there was an overall confirmation of the prediction, but the two RNAi lines tested for one given target did not share the same effect or we were able to test only one line. The largest group (19/51) was made by cases in which there was no effect and surprisingly in a remarkable number of cases (14/51) there was an overall effect opposite to that predicted, albeit either the two RNAi lines tested for one given target did not share the same effect or we were able to test only one line.
In addition to the 2-RNAi lines criterion we have devised a quantitative index for ranking these targets by combining their effect in the RNAi screen (averaging the Chi square for the RNAi lines targeting each gene, Av(χ2)IR) with the strength of the prediction in all combined databases (Σ{Normalised Σ[(Score)*Av(χ2)]}).
This parameter (Σ{Normalised Σ[(Score)*Av(χ2)]}*{Av(χ2)IR}) highlighted garz, one of the six targets satisfying the 2-RNAi lines criterion, as the top target (Table 3). However, there was incomplete agreement with respect to the rest of the ranking between the two criteria, i.e. our scoring system and the rule of 2-RNAi lines, with only four of the ten top scores coming from target genes satisfying the 2-RNAi lines criterion.
We also tested 14 additional targets that were differentially predicted by the different databases. We were able to further identify five targets that confirmed the predicted phenotype, one satisfying also the 2-RNAi lines criterion, while two had the opposite overall effect (Table 4).
A comparison between these two groups, the common to all databases and the differentially predicted, highlights that the fraction of validated prediction is similar, but the chance of finding false positives (i.e. targets that had the opposite effect to that predicted) is paradoxically higher in the commonly predicted group (15/51 in the common and 2/14 in the differential).
Considering the lack of tangible benefits of focusing on the commonalities between the different databases, we have then exploited our validation analysis to quantify the prediction capability of each of the four databases to identify the most valid for our screen. For all targets tested, both from the common group (Table 3) and from the differential group (Table 4), we have calculated the database-specific Normalised Σ[(Score)*Av(χ2)] *{Av(χ2)IR} parameter by combining the quantification of the lifespan effect of the RNAi lines (average Chi square in the RNAi screen) with the normalised predicted score from each database. Then, to rank databases we have summed all these results (with a negative value for false positives) to determine the predicting power score. TargetScan had the highest predicting power for the list of common targets, while MicroCosm had the highest capacity for target identification among the differential targets. PicTar had the lowest predicting power in all cases. However, MicroCosm also predicted the largest number of genes as targets of our miRNA screen, with over 44% of them not shared by the other databases. We reasoned that this lack of efficiency in EBI Microcosm had to be considered and when normalising for the total number of predicted targets from each database, as a measure of the predicting power efficiency, TargetScan showed a greater efficiency in both cases, followed by miRNA.org.
As mentioned, we ranked the target genes from the RNAi confirmed predictions and decided to further investigate the top ranked target, garz, the fly orthologue for GBF119,20,39).
Pan-glial knockdown of garz with repo-Gal4 specifically during adulthood strongly reduced lifespan. This was true for both RNAi lines tested when compared to w1118 median lifespan control (Figure 1B). Different glial cell types present in the adult fly brain have specific morphology and function40. In order to test if a specific glial sub-population could account for the observed phenotype, we targeted the knockdown of garz using established Gal4 driver lines: astrocyte-like (alrm-Gal4), cortex (NP2222-Gal4), subperineural (moody-Gal4) and peripheral (gliotactin-Gal4) glia. In all sup-populations tested, the downregulation of garz caused a reduction in lifespan, albeit not as strong as the pan-glial knockdown (Figure 1C). This suggests that a combination of multiple functions is affected by garz.
We also analysed the effects of pan-neuronal (elav-Gal4) knockdown of garz. This also led to a significant shortening of lifespan although the effect was milder than the one obtained with pan-glial garz knockdown (Figure 1B vs 1C), either because of differences in Gal4 line strength or because of a higher impact of garz function in glial cells for maintenance of the brain homeostasis.
We then focused on rescuing the glia-related shorter lifespan phenotype using exogenous transgenes for garz and human GBF1. Although the overexpression of garz alone in adult glia had a very toxic effect, when combined with the garz-RNAi overexpression, promoted a modest but significant rescue of the lifespan (Figure 1D). This suggests that garz levels need to be tightly controlled in the fly. On the other hand, overexpression of the human GBF1 was entirely neutral for fly lifespan when expressed on its own and fully rescued the lifespan phenotype when co-expressed with garz RNAi. This indicates a remarkable conservation in functions between garz and GBF1.
For both garz and GBF1, the presence of a functional Sec7 domain, which is responsible for the catalytic activity of GEF proteins domain24, was important to exercise their rescue activity (Figure 1D). In the case of UAS-garz, a mutation of the Sec7 domain entirely eliminated the rescue of garz knock down, actually aggravating toxicity. This also indicates that the toxicity of garz overexpression is not dependent on the catalytic GEF function of garz, possibly suggesting a dominant negative effect by sequestration of binding partners in catalytically inactive complexes. Additionally, in the case of UAS-GBF1, the rescue effect was significantly reduced, albeit not entirely eliminated, by an inactive Sec7 domain (Figure 1D).
Human GBF1 showed a remarkable capability to fully rescue lifespan shortening upon garz knockdown in glia. We then asked whether it would also be able to rescue the lifespan shortening induced by miRNAs predicted to target garz. From our database analysis, miR-1, miR-79 and miR-315, all causing a strong reduction of lifespan11, were among the miRNAs predicted to target garz and may be rescued by GBF1. Indeed, UAS-GBF1 was able to significantly rescue the phenotypes caused by the overexpression of these miRNAs in glia (Figure 1E). GBF1 co-overexpression was able to rescue the lifespan for miR-79 and miR-315 to what would be commonly observed in wild-type flies. These results confirm our initial predictions and establish garz as the main mediator of the effect on lifespan caused by overexpression of miR-79 and mir-315 in adult glia. The partial rescue of the miR-1 phenotype indicates that garz is only partially responsible for the effect of miR-1 in adult glia and is in accordance with the previously reported role of repo in miR-1-mediated lifespan shortening11.
We have previously described an automated unbiased and high-throughput method to analyse fly motor activity11). When using this paradigm, we unravelled an impact of glial garz knockdown on the amplitude of the response to a train of stimuli and GBF1 co-overexpression rescued this response (Figure 2A). When looking at spontaneous activity parameters, i.e. non-stimulus driven, in the same experiment, flies expressing garz-RNAi showed a reduced average speed and an increased interval between bouts of movement without reflecting in the overall bout movement duration. Both average speed and inter-bout interval were fully rescued by the co-overexpression of GBF1 (Figure 2B–D). This analysis indicates that garz knock down affects not only lifespan but also the healthspan and motor activity both exogenously stimulated and internally generated, making flies slower and also pausing more.
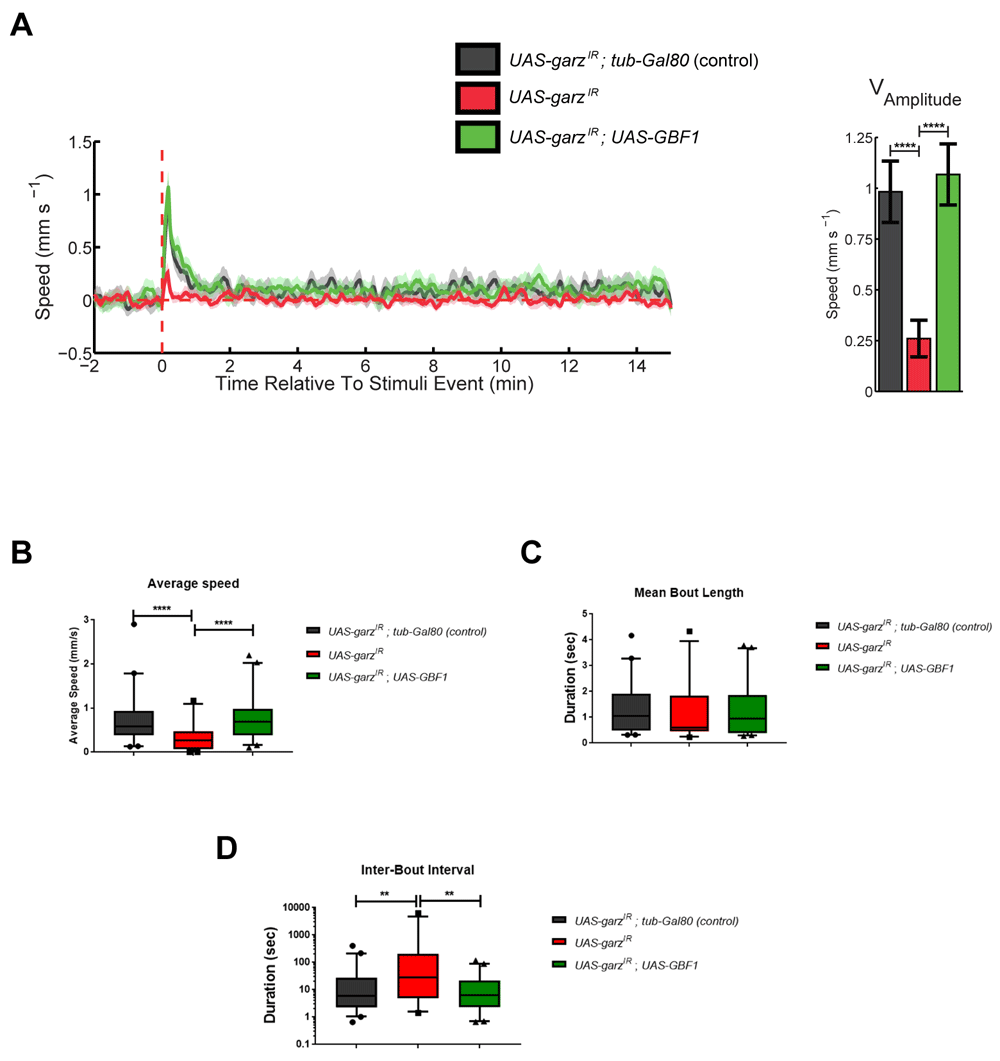
All data in this figure represent a grouping of two independent experiments with a total number of flies analysed (N) of 35–40. Error bars represent SEM in all graphs. Untreated track data can be accessed at DOI 10.17605/OSF.IO/UNJX7. (A) Stimulus response curve for control flies (black), garz RNAi (red) and co-expression of GBF1 and garz RNAi (green). The graph is an average of 6 tracks for each of the stimuli received at 15 min intervals (See Methods). All genotypes also include repo-Gal4 and ubi-Gal80ts to express the transgenes in all adult glia. In control flies the presence of tub-Gal80 blocks any expression of UAS-transgenes. The graph to the right reports the mean amplitude of the response to a train of stimuli, which is significantly reduced by RNAi against garz, and this reduction is reverted to normal level by co-expression of human GBF1. One-way ANOVA, Dunnett’s multiple comparisons post hoc test. (B) Average speed analysis of the same flies as in A. RNAi against garz significantly slows down fly motility and this is rescued by human GBF1. One-way ANOVA, Dunnett’s multiple comparisons post hoc test. (C) Mean bout length analysis of the same flies as in A. No significant difference is detected in this parameter. One-way ANOVA, Dunnett’s multiple comparisons post hoc test. (D) Mean interbout interval analysis of the same flies as in A. RNAi against garz significantly increases the time spent in inactivity by flies and this is rescued by human GBF1. One-way ANOVA, Dunnett’s multiple comparisons post hoc test.
We next set out to determine the effects garz knock down had inside the glial cells that would correlate with behavioural and lifespan dysfunctions.
It has been reported that garz knockdown impairs vesicle transport and membrane delivery during fly development25. Thus, we analysed membrane distribution in the presence of garz-RNAi in adult brains. Driving the expression CD8-GFP in glia showed aberrant membrane distribution upon garz knockdown when compared to a more homogeneous distribution of the GFP signal in glia from control brains (Figure3A, Videos 1 and 2). Such data suggests that overall membrane trafficking in glia may be impaired although we have not been able to detect failure in membrane delivery of the cell adhesion cadherin molecule CadN, whose potential glia localisation effects may, however, be masked by the unaffected CadN localisation in neurons, where CadN is highly expressed (data not shown, the full dataset can be accessed at DOI 10.17605/OSF.IO/7HRZS).
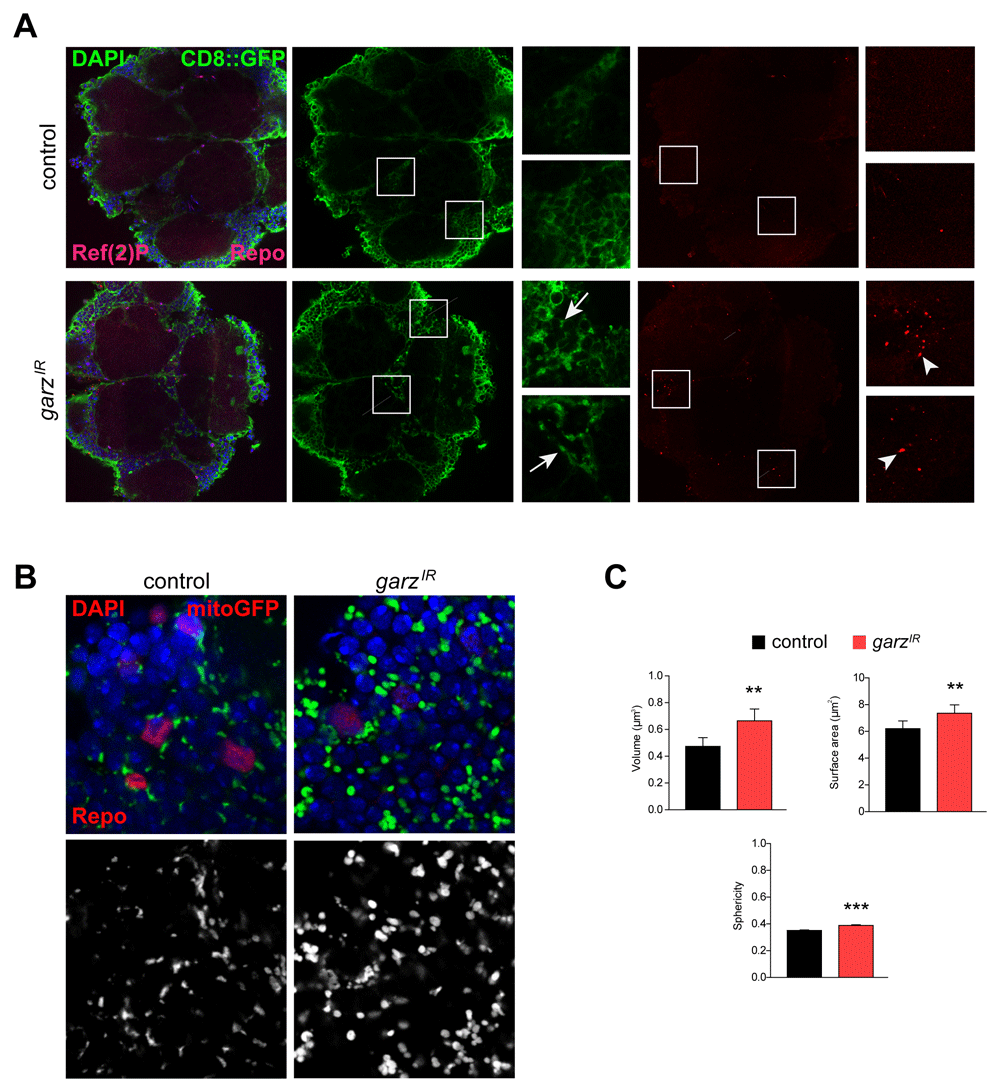
(A) Representative single confocal sections of adult fly brains stained for DAPI (blue), GFP (green), Repo (magenta) and Ref (2)P (red). Pan glial knock-down of garz with repo-Gal4 and ubi-Gal80ts leads to abnormal distribution of the plasma membrane targeted CD8-GFP protein (expressed from a UAS-CD8-GFP transgene in all glial cells) leading to gaps and blebs (arrows, see also Videao1 and 2), and to accumulation of Ref(2)P puncta (arrowheads). The full dataset can be accessed at DOI 10.17605/OSF.IO/96TS3. (B) Representative single confocal section of adult fly brains stained for DAPI (blue), GFP (green) and Repo (red). The GFP signal also in back and white (lower panels) is due to the presence of a UAS-mitoGFP transgenes and detects mitochondria. (C) Quantification of mitochondria parameters based on the GFP signal in B. Pan glial knock-down of garz with repo-Gal4 and ubi-Gal80ts leads to significant increases in the volume, surface area and sphericity of mitochondria. Mann-Whitney non-parametric test. N=300 objects, randomly selected from 4 brains. Error bars represent SEM. The full dataset can be accessed at DOI 10.17605/OSF.IO/EXMTG.
Conflicting in vitro data has been reported for the effects of GFB131 and garz32 in what concerns autophagy regulation. Looking at the distribution of the Ref(2)p (the orthologue of mammalian p62) autophagy receptor43 revealed Ref(2)p accumulation in puncta, suggesting a potential block in autophagic clearance in glial cells (Figure 3A).
Finally, it has been suggested a role for GBF1 in the regulation of mitochondria morphology and function in yeast, C. elegans muscle and HeLa cells30. Using mito-GFP transgene we were able to identify mitochondrial morphology defects in adult glial cells (Figure 3B, C). Quantification of the main morphological parameters has unravelled an overall increased mitochondrial volume, surface and sphericity upon garz knockdown. These parameters may indicate a defect in mitochondria quality control and are in agreement with an impaired autophagic clearance, which has the potential to also affect mitophagy.
Underlying data contains the raw data behind these results44.
We have previously screened a library of miRNAs for effects on Drosophila’s lifespan when expressed in adult glia and already established that this strategy can identify factors important for nervous system health in adult life11. We aimed here at developing a generalizable global approach that would allow to identify the key target genes that mediate the actions of miRNAs in a given context. Focusing on miRNAs that shortened the lifespan, we devised an in-silico strategy to unravel a potential list of genes relevant for glial function and consequently brain homeostasis in adult flies. The outcome of this strategy had efficiency issues and highlighted the little overlap in the predictions made on the basis of four different databases for miRNA target prediction in Drosophila.
To put to the test the outcome of these in silico predictions, we have silenced individual genes by inducing the expression of specific RNAi in adult glia. The assumption being that RNAi downregulation of the top target genes would phenocopy the effect observed when expressing the miRNAs targeting them, i.e. lifespan reduction. Overall, however, the number of genes that, upon knockdown, reduced lifespan was remarkably low, and we could observe no tangible benefit of focusing on predictions in common to all four databases, versus targets differentially predicted in the different databases. It was also evident from our analysis that, among the databases, TargetScan and miRNA.org were considerably more efficient in delivering predictions that withstood the RNAi tests.
Therefore, the benefits of using miRNAs-based screens and in silico identification of targets, in place of much larger screens based on targeting single genes, have to be carefully evaluated and in silico selection of target genes should be based primarily on the TargetScan and miRNA.org databases. Nevertheless, the fraction of validated positive target genes by two criteria (7/65) and by at least one (22/65) is much larger than what usually expected in siRNA screens and suggests a 3/5-fold enrichment in positive hits. Thus, our method makes Drosophila screens a more appealing platform with reduced workload in comparison to traditional single gene targeted screens, whether by RNAi or genomic mutagenesis. This may have 3Rs benefits, facilitating the use of Drosophila as a model for preliminary studies on the genetic factors that influence a given biomedical process.
Our screen has also highlighted a number of genes that are strong, and in most cases unexpected, candidates for essential functions in adult glia in ageing. This list of genes provides a useful tool for scientists studying glial functions in ageing. In particular, all identified genes that have been validated by two RNAi lines have clear mammalian orthologues. Drosophila can therefore be used to study in detail the functions of these genes in the adult glial cells, in place of genetically modified mouse models.
To validate our findings, we focused on the top target of the genes commonly predicted by all databases and also by TargetScan, i.e. garz, the fly orthologue of the human GBF1.
The analysis of garz confirmed that this gene is absolutely required in adult glia, and also in neurons, for fly survival. Using our automated behavioural set up we could also establish that garz is essential in glia for locomotor activity in response to a stimulus or endogenously generated. Analysing the effects of silencing garz in different glial sub-populations showed that the strong reduction in lifespan could not be accounted for by one specific type of glia but rather due to a combined effect of silencing garz in all glial cells simultaneously, indicating that garz is essential for any glial cell type.
Our subcellular analysis suggests that the locomotor and lifespan defects correlate and possibly originate from a number of cellular defects in protein trafficking, autophagy and mitochondria quality control.
In Drosophila, mutated versions or knockdown of garz resulted in developmental epithelial morphogenesis defects20,21 and impaired membrane delivery of adhesion molecules25. We have been able to identify membrane defects in glial membrane distribution, although not all membrane proteins seemed to be affected by garz knockdown. garz and GBF1 have been identified as a positive autophagy regulator in Drosophila primary cultured muscle cells32 and mammalian cells31. An accumulation of Ref(2)P upon garz-RNAi expression in adult glia suggests an autophagic clearance deficits, in agreement with these studies.
GBF1-RNAi has been shown to affect mitochondrial morphology and function30. Chemical inhibition of GBF1 in mammalian cells also showed condensed mitochondria and mislocalisation in the cell45. Although mislocalisation of mitochondria is difficult to assess due to glial cell morphology in the Drosophila brain, garz-RNAi strongly affected mitochondria morphology suggesting a more condensed state which may be a reflection of an unbalanced fission/fusion regulation and mitochondria quality control46.
Our analysis further suggested that there was remarkable functional conservation between garz and human GBF1. While the lack of toxicity of GBF1 overexpression, in comparison to Garz, may indicate some divergence and lack of dominant negative activity, this may also be due to different levels of expression or tags. Nevertheless, GBF1 was able to fully rescue, partially in a Sec7-domain dependent manner, the shorter lifespan and motor behaviour phenotypes caused by the silencing of garz. GBF1 was also able to rescue the lifespan shortening by three different miRNAs, miR-1, miR-79 and miR-315, validating that in our screen their effect is at least partially, and in some cases almost entirely, due to downregulation of garz.
Thus, these data validate both the logic and principles of miRNA screens, despite inefficiencies, and the use of Drosophila as a valid organism to study the biology of garz/GBF1.
The identification of major cellular events regulated by garz/GBF127–29,47–49 has targeted such molecules for health and disease studies18. Recently, it has been shown that siRNA knockdown of GBF1 causes intracellular Amyloid Precursor Protein (APP) accumulation in primary cortical neurons; overexpression of GBF1 contributes to APP trafficking and is dependent on its GEF activity50. Inhibition of GBF1 with brefeldin A was also shown to lead to a new form of cellular degeneration and death in neurodegenerative diseases, based on destruction of the nuclear lamina51.
Gbf1 conditional mutant mice have been generated in the Wellcome Trust Sanger Institute and are being phenotyped by the International Mouse Phenotyping Consortium (https://www.mousephenotype.org/data/genes/MGI:1861607). We demonstrate here that Drosophila would constitute an ideal organism to put forward 3Rs-compliant alternatives and, at least partially, replace this mouse line in studies aiming at understanding the role of GBF1 in health and disease.
Open Science Framework: miRNA-garz. https://doi.org/10.17605/OSF.IO/A5ZST44.
This project contains the following underlying data:
Table 2 Data – Pimental et al., 2020 (XLSX). (Data underlying Table 2.)
Table 3 Data – Pimental et al., 2020 (XLSX). (Data underlying Table 3.)
Table 4 Data – Pimental et al., 2020 (XLSX). (Data underlying Table 4.)
Figure 1C Data - Pimentel et al., 2020 (XLSX). (Data underlying Figure 1C.)
Figure 1D Data - Pimentel et al., 2020 (XLSX). (Data underlying Figure 1D.)
Figure 1E Data - Pimentel et al., 2020 (XLSX). (Data underlying Figure 1E.)
Figure 2 Data - Pimentel et al., 2020 (XLSX). (Data underlying Figure 2.)
Figure 3A and videos. (TIFF images and ZIP files containing data underlying Figure 3A.)
Figure 3B-C. (ZIP files containing raw images underlying Figure 3B, C.)
Extended data Table 1- Data - Pimentel et al., 2020 (XLSX). (Data underlying Extended data Table 1.)
Extended data Table 2- Data - Pimentel et al., 2020 (XLSX). (Data underlying Extended data Table 2.)
Extended data Table 3- Data - Pimentel et al., 2020 (XLSX). (Data underlying Extended data Table 3.)
Extended data Table 4- Data - Pimentel et al., 2020 (XLSX). (Data underlying Extended data Table 4.)
Data not shown. (ZIP files containing images of membrane delivery of the cell adhesion cadherin molecule CadN.)
Open Science Framework: miRNA-garz. https://doi.org/10.17605/OSF.IO/K5HW938.
This project contains the following extended data:
Extended Data Table 1. MicroCosm target prediction and ranking tables. For each miRNA, ranking of target prediction - column (Score)*Av(χ2) - was made by multiplying the Average χ2 obtained in the screen (from Table 1) by the Score predicted in the MicroCosm database. In the total table, all values from a given target, resulting from all miRNAs were summed in a final ranking value in column Σ(Score)*Av(χ2)]. This table and the full dataset can be accessed at DOI 10.17605/OSF.IO/R3ZX9.
Extended data Table 2. PicTar target prediction and ranking tables. For each miRNA, ranking of target prediction - column (Score)*Av(χ2) - was made by multiplying the Average χ2 obtained in the screen (from Table 1) by the Score predicted in the PicTar database. In the total table, all values from a given target, resulting from all miRNAs were summed in a final ranking value in column Σ(Score)*Av(χ2)]. This table and the full dataset can be accessed at DOI 10.17605/OSF.IO/MDKHR.
Extended data Table 3. miRNA.org target prediction and ranking tables. For each miRNA, ranking of target prediction - column (Score2)*Av(χ2) - was made by multiplying the Average χ2 obtained in the screen (from Table 1) by the square value of Score predicted in the miRNA.org database. The square value was used in this case as the scoring system used by miRNA.org delivers negative values, differently from the other databases. In the total table, all values from a given target, resulting from all miRNAs were summed in a final ranking value in column Σ(Score2)*Av(χ2)]. This table and the full dataset can be accessed at DOI 10.17605/OSF.IO/539J8.
Extended data Table 4. TargetScan target prediction and ranking tables. The TargetScan database does not provide a scoring system for its predictions, rather a list of 8mer or 7mer sequences matched by the miRNA on the target and an information on the conservation of these sequences. We have attributed a numerical score to these sequences privileging the importance of 8mer vs 7mer and of conservation according to the scheme described in the Methods section. For each miRNA, ranking of target prediction - column (Score)*Av(χ2) - was made by multiplying the Average χ2 obtained in the screen (from Table 1, some values specifically generated averaging all miRNA grouped in a single family by TargetScan) by the Score obtained according to our above-mentioned scheme. In the total table, all values from a given target, resulting from all miRNAs were summed in a final ranking value in column Σ(Score)*Av(χ2)]. This table and the full dataset can be accessed at DOI 10.17605/OSF.IO/WD6ZR.
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
We thank M. Freeman, E.C. Lai, R. Sousa-Nunes, J. Bateman, S. Luschnig, the VDRC, the DSHB and the BDSC for fly stocks and reagents.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are a suitable application and appropriate end-users identified?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are the 3Rs implications of the work described accurately?
Yes
Is the rationale for developing the new method (or application) clearly explained?
Yes
Is the description of the method technically sound?
Yes
Are sufficient details provided to allow replication of the method development and its use by others?
Yes
If any results are presented, are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions about the method and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Invertebrate models, genetics, neuroscience. Referee suggested by the NC3Rs for their scientific expertise and experience in assessing 3Rs impact.
Are a suitable application and appropriate end-users identified?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are the 3Rs implications of the work described accurately?
Yes
Is the rationale for developing the new method (or application) clearly explained?
Yes
Is the description of the method technically sound?
Yes
Are sufficient details provided to allow replication of the method development and its use by others?
Yes
If any results are presented, are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions about the method and its performance adequately supported by the findings presented in the article?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: ageing, mTOR, autophagy, Drosophila
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 23 Jul 20 |
||
|
Version 1 01 May 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
Please enter your details to receive updates from the NC3Rs
We'll update you about the NC3Rs and the NC3Rs Gateway.
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)