Keywords
Mutational Signatures, pmsignature, COSMIC, Web interface, Shiny, R
This article is included in the RPackage gateway.
Mutational Signatures, pmsignature, COSMIC, Web interface, Shiny, R
We thank the reviewers for their insightful comments. Two major changes have been made to the paper which we believe have improved it significantly.
The first change is that we updated the version of COSMIC signatures from version 3 to version 3.1, announced in June 2020 as the most recently released signatures. The other change was made based on reviewer 2’s comment on including another conversion method. Reviewer 2 suggested that we ‘collapse’ the COSMIC signature to marginal probabilities which are then multiplied together under the independence assumption before comparing the COSMIC to PM signature. We implemented the new method in the Shiny app, introduced it in the Methods section and provided new Results. Now users are able to choose either of these conversion methods (new ‘collapse’ or original ‘expand’) to identify the most similar signature of the opposite type. In addition, a new tab featuring heatmaps was implemented to provide an interactive visualization of the cosine similarity between two types of signatures. Cosine similarities are computed after converting one of the signature types to match the format of the other. In addition, we discussed the discrepancy that can arise in identifying the most similar signature of the opposite type, depending on which conversion method is selected (‘collapse’ or ‘expand’).
Based on the reviewers’ feedback, we have also made a few minor changes including 1) adding a new figure to illustrate how to convert between two types of signatures; 2) correcting the typos in the formula, text, and Shiny app user interface; 3) updating a reference and the Shiny app user interface accordingly.
See the authors' detailed response to the review by Adrian Baez-Ortega
See the authors' detailed response to the review by Vittorio Perduca
Each human is subject to a variety of mutational processes throughout their lifetime. These processes result in a catalog of somatic mutations in the tissue creating a unique mutational profile1. A mutational signature captures the pattern of the mutations and contexts in which those mutations occur (i.e., the neighboring bases). Examples of important mutational processes with distinct mutational signatures include aging and ultraviolet (UV) radiation. Additionally, many research groups are performing analysis to discover de novo mutational signatures in cancer1–4.
Currently, there are two frameworks used to characterize and visualize mutational signatures5,6. The first, proposed by Alexandrov et al., uses a vector of 96 probabilities to capture the composition of the six nucleotide substitutions (C >A, C >T, C >G, T >A, T >C, T >G) and the neighboring base immediately on each of the 5′ and 3′ side of the mutated base1. A list of published mutational signatures can be downloaded from the Catalogue Of Somatic Mutations In Cancer (COSMIC) website7 (version 2, v2). Later, Alexandrov et al. published an expanded set of mutational signatures in version 3.1 (v3.1)8. The 72 COSMIC v3.1 Single Base Substitution (SBS) signatures include 30 v2 signatures. Based on the signature concept, but using different model assumptions, Shiraishi et al. proposed a mixed-membership model, pmsignature, which substantially reduced the number of parameters needed to characterize a signature9. They achieved this by assuming independence across bases, thereby reducing the number of parameters from 6*4*4-1 = 95 to (6-1)+(4-1)+(4-1) = 119. The reduction in the number of parameters is greater if more flanking bases are included. However, the independence assumption might prevent signatures with dependent neighboring bases from being discovered, thereby resulting a fewer signatures. Shiraishi identified 27 signatures, all of which can be downloaded from their GitHub repository9. In this paper, we will refer to signatures resulting from these two methods as “COSMIC signatures” with version numbers (for those resulting from Alexandrov et al.’s method) and “PM signatures” (for those resulting from Shiraishi et al.’s method).
A large number of researchers have published scientific findings resulting from the COSMIC signature-based method10–12, which was defined as the “gold standard" in the field by Baez-Ortega et al.6. Meanwhile, an increasing number of researchers are using the pmsignature-based method for samples with lower numbers of somatic variants due to it requiring fewer parameters9,13,14. Given that both methods are widely used, investigators need the ability to compare results from their analysis with those reported in earlier databases, which may have been produced using the alternate method. For example, researchers have adopted both tools for gastric cancer and tried to compare and integrate the information from two data sources in a somewhat ad hoc manner15. No rigorous tool exists for this task. In this paper we present iMutSig, an easy-to-use tool that allows users to 1) input a new mutational signature, 2) compare it using cosine similarity to all published signatures from both the COSMIC and PM signature databases, 3) identify the most similar signatures previously reported, and 4) to assemble the information characterizing those signatures using simple point-and-click navigation.
In order to measure the similarity between mutational signatures across two databases, we need to represent PM signatures in a way that is comparable with those from COSMIC, or represent COSMIC signatures in a way comparable to PM signatures. We call the first of these methods the “expand” method, where we expand the PM signature into a probabilistic vector with the same length as the COSMIC signature, i.e., 96. The conversion in the opposite direction, from the COSMIC signature into the PM signature format is called the “collapse” method. In the collapsed format, the PM signature is represented by a vector of 14 probabilities, the probabilities for the six possible nucleotide substitutions and the probabilities for the four possible bases at each of the two flanking base positions. In the “expand” method, to calculate each of 96 resulting probabilities in the vector, we take the constituent components that make up the COSMIC signature - which refer to the nucleotide substitution and two flanking bases at the -1 and +1 position - calculate the probability of each component for the given PM signature, and then multiply those probabilities using PM signature’s assumption of independence. For example, to calculate the probability of the COSMIC signature C[C >A]T we multiply three PM signature’s probabilities: P(C at pos -1), P(C >A), and P(T at pos +1). This example is shown in Table 1, Equation 1, and Figure 1.
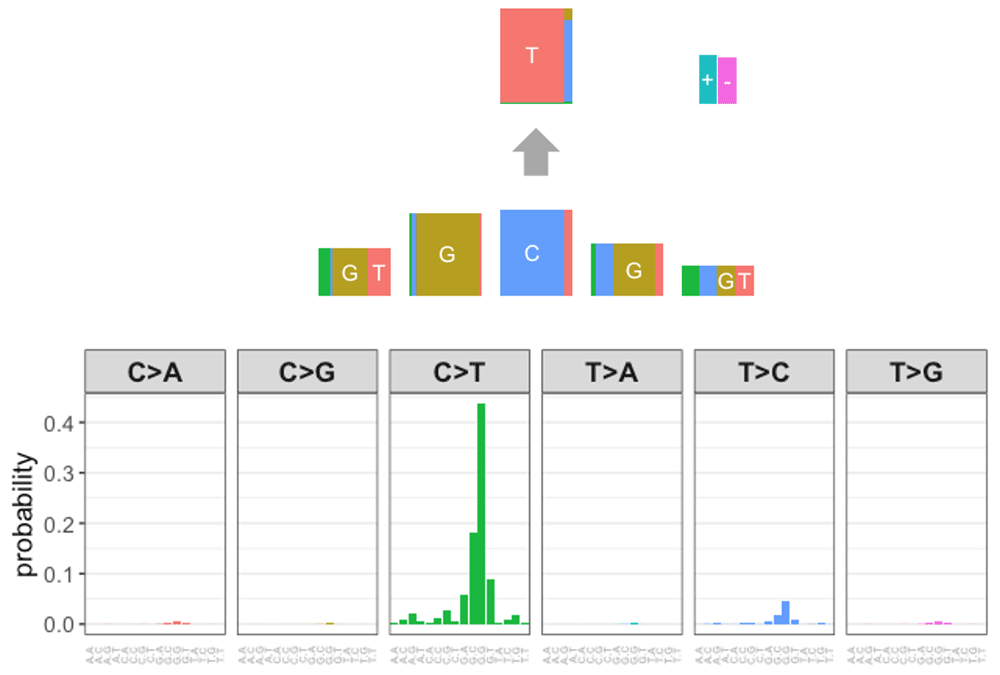
To perform the “collapse” method, we calculate the marginal probability for each characteristic, the nucleotide substitution and each flanking base, and multiply the probabilities together using the independence assumption. The marginal probability for the nucleotide substitution is computed by summing the probabilities including all 16 combinations of two flanking bases from the COSMIC signature. In a similar manner, the marginal probability of a flanking base is the sum of probabilities across all signatures containing the given flanking base. See an example of P(C>A) and P(C at pos -1) shown in Equation 2:
These are computed using the convertAlexandrov2Shiraishi function from the decompTumor2Sig package15.
After we have represented both forms of signature using probabilistic vectors of the same length n, P and C say, we can directly compare the two signature types. In order to measure the similarity between them we use cosine similarity, CS, defined as shown in Equation 3:
Intuitively speaking, cosine similarity is the cosine of the angle between the two vectors. As such, cosine similarity ranges from 0 to 1 (inclusive). In our context, if two mutational signatures have a cosine similarity of 1, they must be identical, i.e., the angle between them is 0°; in contrast, if two mutational signatures have a cosine similarity of 0, they are maximally dissimilar (i.e., orthogonal). Computing the cosine similarity between the input signature and each of the candidate signatures, and then sorting the similarities from highest to lowest value, we identify the candidate signature with the highest cosine similarity as the most similar mutational signature.
iMutSig is built in R with its key features depending on the R package, pmsignature9. As shown in Figure 2, the Shiny app currently supports three possible workflows for users to choose from, depending on the type of signatures they have already obtained: 1) starting with a COSMIC signature; 2) starting with a PM signature; 3) starting with a self-defined signature that could follow either the COSMIC or PM format.
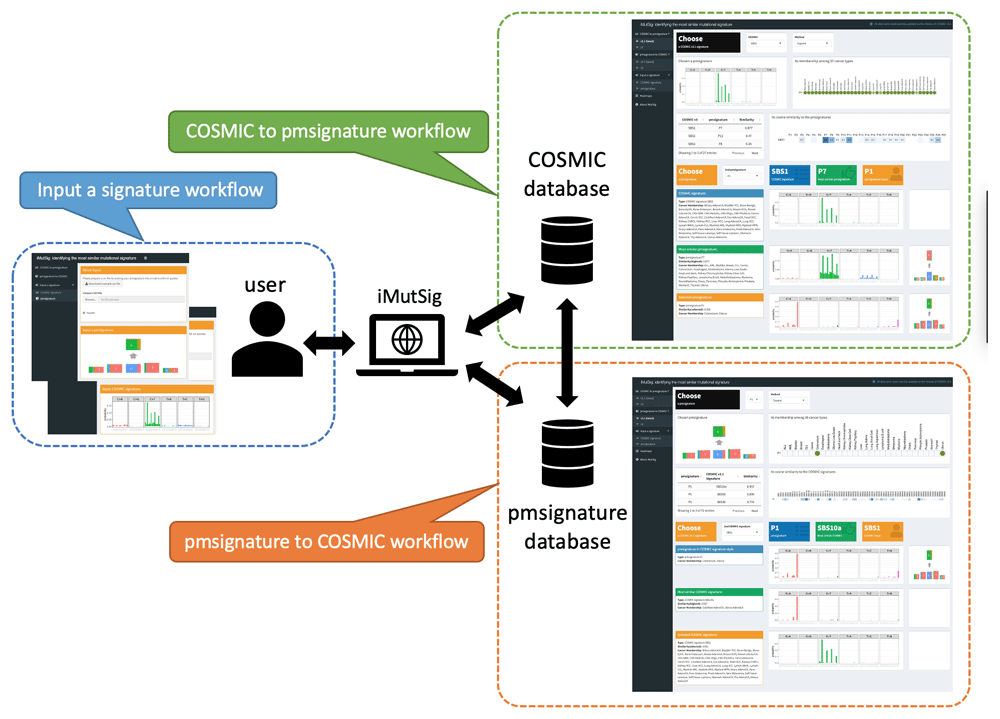
The first two tabs allow users to finding the most similar PM signature to an input COSMIC signature (highlighted in green) and vice versa (highlighted in orange). In addition, users can identify the most similar signatures from both data sources to an input signature (highlighted in blue).
The first tab in the Shiny app window, “COSMIC to pmsignature", allows users to select an input COSMIC signature via a drop-down list and returns the best-matched PM signature. The returned results are divided and organized separately in the top and the bottom portion of the page. The top half tab summarizes background information regarding the input signature by presenting: 1) visualized plots of the input signature and its membership among all cancer types, i.e., in which kind of cancers the mutational signatures has been found; 2) a table showing the cosine similarity between this signature and all PM signatures, sorted in decreasing order, along with a visualization of a similarity heatmap with color and intensity proportional to assessed similarity. The bottom half tab presents plots and descriptions of the input COSMIC signature, the most similar PM signature, and a second PM signature that the user can select. Thus, users can easily access all the vital information and results regarding these signatures rather than having to manually gather and organize information from publications. The top half of the tab will be automatically updated via a control panel in the middle section of the tab, which enables users to select a signature to start with and also highlights information about the currently selected signature, the most-similar signature from the alternate model framework, and the cosine similarity.
The second tab was designed in a similar manner to the first tab, but for the case in which we are starting with a PM signature and looking for the most similar COSMIC signature. For the first two tabs, users can choose which version of COSMIC signatures to input from the sub-menus, i.e., v2 or v3.1.
Unlike the first two tabs, the third tab enables users to enter a user-supplied signature, which can be in either PM or COSMIC format, and then identify the most similar signature from each online database. The user will be requested to enter a sub-menu based on the type of the input signature and to upload a comma-separated values (CSV) file containing a single signature. A sample CSV file is provided for download to give the user a better sense of the format of the input file. Then, the tab will be updated to display three tables, one from each data source (COSMIC v2, v3.1 and PM), listing the signatures from that data source and the cosine similarity of each signature with the user-uploaded signature. The tables are ordered from most similar to least similar signature. In addition, the user is able to view figures of the best-matched signatures (i.e., those with highest cosine similarity) from each data source, allowing users to observe any similarities and dissimilarities. Below, users will see a list of cancer types that contain the best-matched signature.
The fourth tab shown in Figure 3 displays the interactive cosine similarity heatmaps between PM signatures and COSMIC signatures for the two conversion methods. One would choose the version of COSMIC signatures (v2 or v3.1) and one of the two conversion methods (COSMIC to PM signature, ‘collapse’, or PM signature to COSMIC, ‘expand’). The PM signature, the COSMIC signature names and the associated cosine similarity value can be visualized by placing the cursor over the heatmap. It is notable that the cosine similarity values tend to be higher using the collapse representation compared to the expand representation. We attribute this to the difference in model assumptions. When a COSMIC signature is collapsed to the PM signature format the independence assumption is imposed on both signature types. However, when a PM signature is expanded to the COSMIC signature format, the PM signature probability vector still represents the fit under feature independence whereas the COSMIC signature does not. This difference in model assumptions results in lower estimates of cosine similarity. Some discrepancies are found, based on the conversion method selected, when searching for the most similar signature from the opposite database: matching COSMIC v3.1 signatures to PM signatures 17 out of 72 disagreed (23.6%). A similar fraction disagreed when matching COSMIC v2 to PM signatures (7 out of 30, 23.3%). Interestingly, when we compare the 27 PM signatures to COSMIC, we see much better agreement with the newer v3.1 signatures compared to the earlier v2 signatures (88.9% vs 63%). The higher matching of the v3.1 database includes the matching of signatures that were not present in the earlier v2 database (e.g. SBS10b, SBS46, SBS49). The remaining discrepant results may correspond to COSMIC signatures that reflect dependence between neighboring bases.
We use iMutSig to identify the most similar signature for a given PM/COSMIC signature or a user-supplied signature. Figure 4 shows the input panel after inputting COSMIC v3.1 signature SBS1 and Figure 5 shows the input panel after inputting PM signature P1. If users provide a user-supplied signature of either COSMIC-kind or PM-kind, the results can be seen in Figure 6 and Figure 7. Consider the example shown in Figure 6, where we input COSMIC v2 signature C1. iMutSig returned the most similar signatures COSMIC v3.1 signature SBS1, and PM signature P7 (similarity = 0.947, and 0.948, respectively) along with the names of its associated cancer types. When providing PM signature P1, iMutSig returned COSMIC v2 signature C10, v3.1 signature C10a and PM signature P1 (similarity = 0.816, 0.957, 1.0, respectively).
iMutSig is a user-friendly interactive browser-based application that allows users who have a signature that they have discovered in an analysis of their own data to identify the best-matched existing mutational signature from the COSMIC and PM databases. It also allows users to directly compare signatures between the two databases. It does this in an interactive way, and also allows straightforward visualization of results. iMutSig enables researchers to easily identify the most similar mutational signature and to easily access characteristic information from both data sources without additional software installation and programming of their own.
All data underlying the results are available as part of the article and no additional source data are required.
Software available from: https://zhiyang.shinyapps.io/iMutSig/
Source code available from: http://www.github.com/USCbiostats/iMutSig
Archived source code at time of publication: https://doi.org/10.5281/zenodo.413241616
License: MIT
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Computational biology, Bioinformatics.
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Applied statistics, biostatistics.
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Partly
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
References
1. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, et al.: Signatures of mutational processes in human cancer.Nature. 2013; 500 (7463): 415-21 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Applied statistics, biostatistics.
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Partly
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Yes
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Yes
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Yes
References
1. Omichessan H, Severi G, Perduca V: Computational tools to detect signatures of mutational processes in DNA from tumours: A review and empirical comparison of performance.PLoS One. 2019; 14 (9): e0221235 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Computational biology, Bioinformatics.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 19 Nov 20 |
read | read |
|
Version 1 10 Jun 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)