Keywords
development workflow, bioinformatics pipeline, quality management, healthcare
This article is included in the Bioinformatics gateway.
This article is included in the Software and Hardware Engineering gateway.
development workflow, bioinformatics pipeline, quality management, healthcare
The importance of best practices for bioinformatics analysis and software have been highlighted by many authors who proposed very valuable guidelines for better reproducibility and traceability (Georgeson et al., 2019; Hamburg & Mogyorodi, 2019; Noble, 2009; Sandve et al., 2013). However, reproducing an analysis is often a challenge (Kim et al., 2018). Therefore, guidelines have to be promoted by the computational labs and bioinformatics core facilities to federate the bioinformaticians across common practices for software development. This is especially essential when the bioinformatics pipeline is used in precision medicine to support the diagnostic and theranostic activities. Indeed, many hospitals worldwide use High-Troughput Sequencing in routine clinical practice to guide the therapeutic decision. This new era of genomic medicine is even promoted in healthcare systems at a large scale within several national initiatives, such as France, USA, UK or Australia (Stark et al., 2019).
This evolution has brought Bioinformatics at the forefront of the healthcare process with the bioinformatics pipeline being a fully integrated component of the clinical decision. Compliance with the healthcare laboratory accreditation standards and regulations is thus required for the development and exploitation of the bioinformatics pipeline. In this context, several authors (Hume et al., 2019; INCa, 2018; Matthijs et al., 2016; Roy et al., 2018) recommended in their guidelines that an appropriate code repository tool should be used to enforce version control. It ensures to track the different releases of the bioinformatics pipeline, their validation and the developers involved in their implementation. This must be integrated in a quality management process with standardized protocols approved for a diagnostic use.
There is a wide ecosystem of tools (see Perez-Riverol et al., 2016; Riesch et al., 2020; Zolkifli et al., 2018) that can support the implementation of such protocols. Among them, we capitalized on git for the version-control system as it became a very popular tool in the software engineering community and GitLab for the repository manager as it can be self-hosted. Both tools offer a large set of very powerful functionalities that can be used and combined in multiple ways thus increasing their usage complexity. It is thus mandatory to formalize through detailed guidelines how to use them on a daily basis for the development and deployment of the bioinformatics pipeline. Therefore, we implemented the set of protocols biogitflow (Kamoun et al., 2020) that describes step-by-step all the command lines and actions to be performed by the developers. A comprehensive documentation available at https://biogitflow.readthedocs.io provides all the technical details. We introduce here the main concepts, steps and principles.
The development workflow consists of four main steps. The first step is software development, which includes code writing and testing. The second step is acceptance testing by the end-users who validate that the expected functionalities have been correctly implemented. The third step is check the installation process and new testing to ensure that the bioinformatics pipeline can be installed in a similar environment than the one used in production. During this step, a new testing is performed such that bugs can be corrected before installing the bioinformatics pipeline in production. Finally, the fourth step is production deployment. During this last step, the new release of the bioinformatics pipeline with the new functionalities is installed in the production environment.
The four different steps have to be performed in separated environments in order to i) ensure that a stable version can be used in production, ii) allow the end-users to validate a new release without any impact on both the version used in production or the version under development and iii) allow the software developers to add new functionalities and modify the code without any impact on the end-users who are validating a new version and/or using the current version in production. Therefore, three deployment environments are used: a development (dev), a validation for pre-production (valid) and a production environment (prod). Besides these environments, each developer can deploy the bioinformatics pipeline in a local workspace to test the new functionalities that have been developed.
Our biogitflow protocols capitalized on the gitflow model proposed by Driessen (2010). The management of the different bioinformatics pipeline versions is based on four different git branches. Depending on the context and the step of the development workflow the following branches on the remote repository are used:
devel contains the code of the current version under development.
release contains the code with both candidate and official releases. The release branch comes from the devel branch.
hotfix is a mirror of the release branch and is used to patch the code that is in production. If a critical bug occurs in production, this branch is used to fix the issue.
master is only used to archive the code from the release and hotfix branches. This branch is not used for development.
Among these four branches, the release, hotfix and master are protected branches such that only the developers with the Maintainer role in the GitLab repository can directly push code on these branches (the other users have to use the GitLab merge request functionality to push on protected branches).
In addition to these four branches, the developer can create local branches to i) implement a new feature (the branch is named with a prefix feature plus any meaningful suffix) and ii) fix a bug in production or resolve a problem during the third step (these branches are either named with the prefix release or hotfix depending on the use case plus some relevant contextual information).
Two levels of roles and permissions are considered. Firstly, in the GitLab remote repository a developer is either assigned to the role Developer (D) to push the developments only on the non-protected branches or Maintainer (M) to push the developments on any branches. Secondly, in the deployment environments a user is either granted with the permission UD to deploy in the dev environment only or UVP to deploy in the valid and prod environments (any user with the UVP permissions is also granted with the UD permissions).
Testing the bioinformatics pipeline occurs during all the steps of the development workflow. It involves all stakeholders including the developers, the users in charge of the deployments and the end-users. This should be done as often as possible to identify and resolve any unexpected behavior early in the development process. The International Software Testing Qualifications Board provides comprehensive guidelines for software testing (Hamburg & Mogyorodi, 2019). Among them, we strongly recommend to include the following testing. The unit testing confirms that a piece of code provides the expected output according to the input parameters. The integration testing checks that the interfaces of the different bioinformatics pipeline components are consistent with each other and that the result of their integration allows the expected functionalities to be performed. The system (or functional) testing validates that the full bioinformatics pipeline works and fits well the end-user’s needs. The regression testing checks that the correction of bugs or the development of new functionalities did not introduce defects in unchanged areas of the bioinformatics pipeline. In addition, we highlight the importance of the operational testing to check that the bioinformatics pipeline provides the expected results on a reference dataset (golden dataset) in the production environment. This testing is systematically launched prior to any new analysis by the bioinformatics pipeline in the production environment. This ensures that the results are reproducible as long as the exact same version of the bioinformatics pipeline is used.
Two use cases have been addressed with dedicated protocols that detail the different actions step-by-step. The first one is the nominal mode (Figure 1) in which a new feature is implemented to improve the bioinformatics pipeline based on requirements and expectations from the end-users who operate it for their daily clinical practice. The second one is the hotfix mode (Figure 2) in which there is a bug in the bioinformatics pipeline in the production environment that hampers the delivery of the results for the patients. The biogitflow documentation provides all the technical details, on how to configure the remote repository in GitLab to develop a new bioinformatics pipeline, how to use git and GitLab depending on the roles and permissions during the time frame of the development workflow for both use cases.
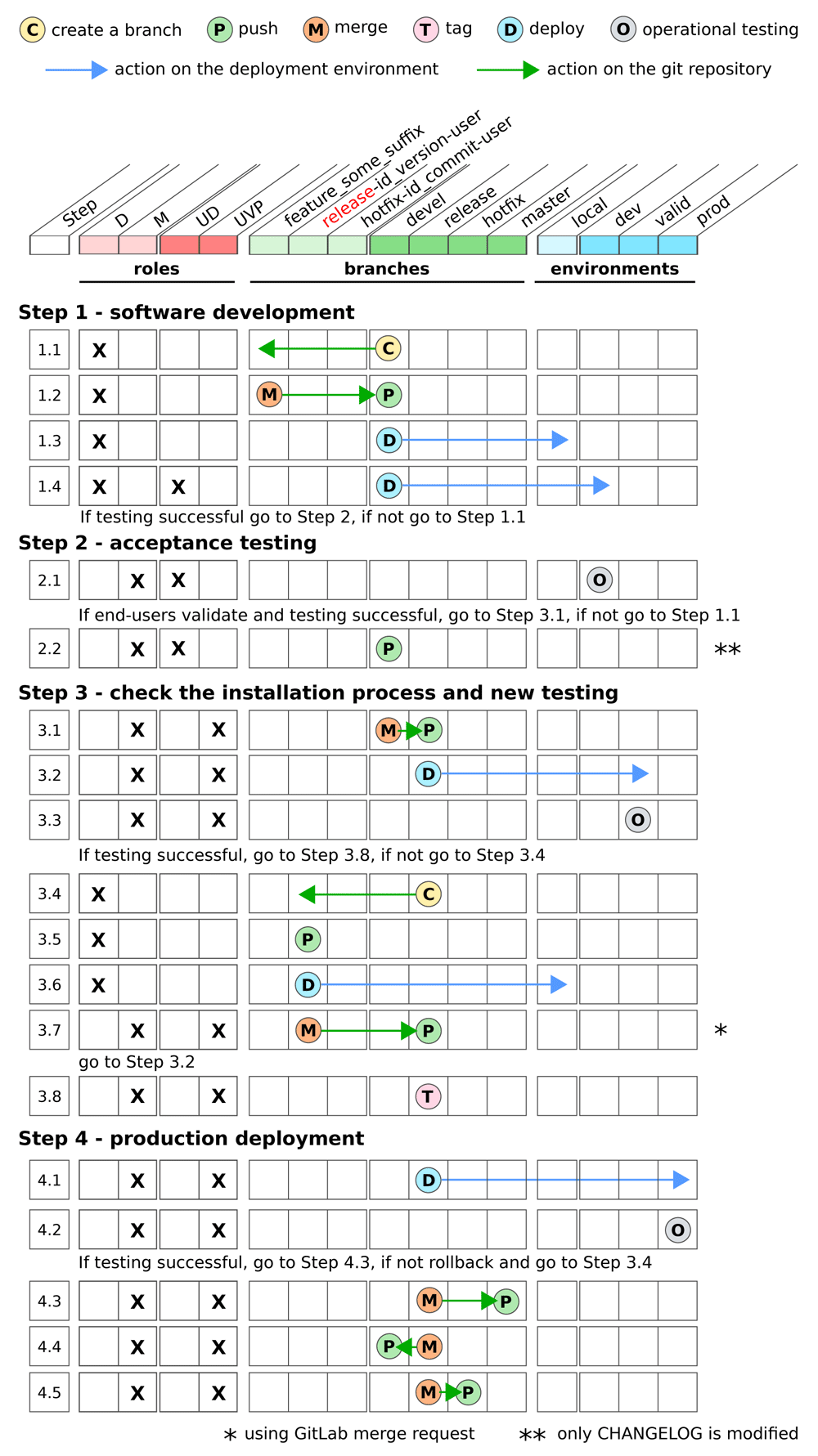
This graphical synopsis provides an overview of the different steps of the development workflow when a new feature is implemented in the bioinformatics pipeline according to the role and permissions of the developer. It describes the different git actions that are performed on the different branches and the deployments in the different environments.
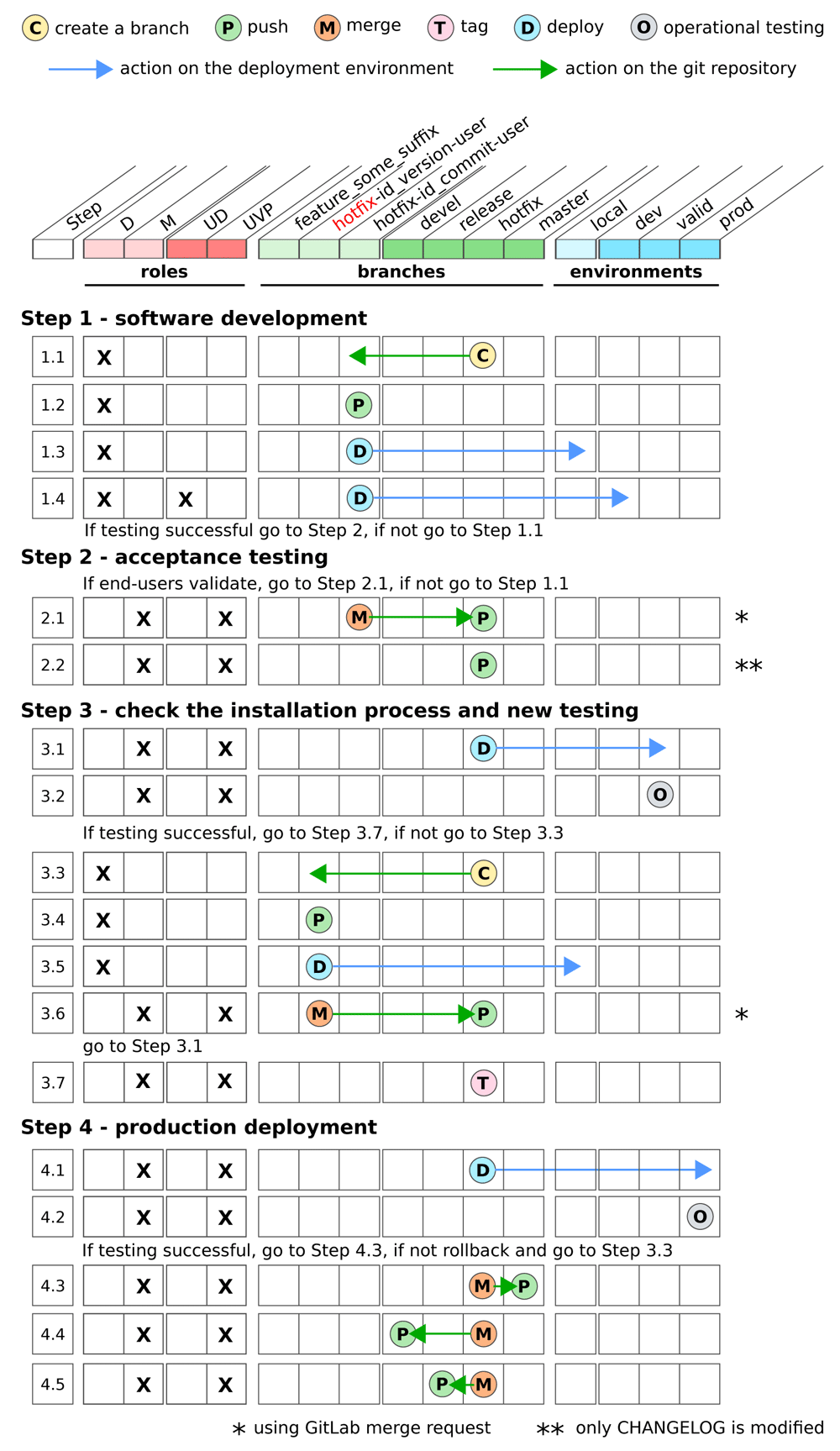
This graphical synopsis provides an overview of the different steps of the development workflow when a bug occurred in the production environment in the bioinformatics pipeline according to the role and permissions of the developer. It describes the different git actions that are performed on the different branches and the deployments in the different environments.
Whatever the use case, the bioinformatics pipeline is deployed in production and operated by the end-users once it has successfully passed all the testing. Whenever new patient data has to be analyzed to deliver results for diagnostic or theranostic purposes, the bioinformatics pipeline can be launched by the end-users only if the operational testing reproduces the results from the golden dataset, otherwise it is blocked by internal control mechanism. In this case, the developers investigate the reason of the failure and have to fix it using the hotfix mode.
As a real example, we provided a biogitflow template of the repository that is available for any registered user at GitLab.com. The user can fork the repository in a personal workspace and apply the nominal mode (Figure 1) and hotfix mode (Figure 2) following the proposed protocoles.
Writing these protocols required some feedback from the developers in order to decide what was the best way to capitalize on git and GitLab taking into account our internal constraints and organization. The need of formalization required by the laboratory accreditation agency such that a bioinformatics pipeline can be used in healthcare was a great catalyst to implement these protocols. It was a major step to improve our daily practice of software engineering towards better quality management and was a source of motivation to change our work habits.
As a corollary of these protocols was the necessity to train the users involved in the development workflow to demonstrate to the laboratory accreditation agency that the technical protocols are known, understood and mastered by the developers. Therefore, an internal accreditation process of our developers was implemented. The training consists of a series of exercises that cover all the different use cases, roles and permissions of the protocols. The exercises are first realized in pair with the tutor and the trainee, then the trainee performs the exercises alone twice, and finally the trainee performs the exercises alone but in the presence of the tutor who ask additional questions to ensure that the tricky parts of the protocols are understood. To be endorsed, the trainee has to perform the exercises fluently in full autonomy. All the actions of the training process are tracked in a dedicated GitLab remote repository. Endorsement is valid for one year that can be extended if the developer still masters the protocols. This is assessed during a dedicated yearly interview with the tutor.
We described two protocols that are used on a daily practice to develop bioinformatics pipelines compliant with the accreditation standards for healthcare. While some choices were made to match our internal constraints and organization, the protocols can be easily transposed in other institutes as the main concepts, steps and principles hold in most of the contexts. According to the principle of the Deming wheel of continuous improvement in quality management, the protocols we described are intended to evolve in order to address future requirements, handle new risk management, integrate new tools or technical frameworks offering better efficiency of the overall process. We strongly encourage the promotion of such protocols not only for the healthcare activities but also for research activities.
All data underlying the results are available as part of the article and no additional source data are required.
biogitflow documentation is available at: https://biogitflow.readthedocs.io/.
Archived source of the biogitflow documentation at time of publication: https://doi.org/10.5281/zenodo.3885463
License: CeCILL Version 2.1.
CK, HDS, JR and PH conceived the protocols. PH wrote the protocols and the manuscript that were reviewed by AG, CK, EG, HDS and JR. PH supervised the study.
We thank the French Bioinformatic Network for NGS Cancer Diagnosis and the Institut National du Cancer (INCA) for the fruitful discussions.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics. Genomics. Cancer. Software Development. Pipelines.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 3 (revision) 19 Feb 21 |
read | |
|
Version 2 (revision) 08 Dec 20 |
read | read |
|
Version 1 22 Jun 20 |
read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)