Keywords
ESBLs, class C β-lactamase, Polymerase Chain Reaction, Ps. aeruginosa, pyocyanin pigment, Khartoum-Sudan.
This article is included in the Pathogens gateway.
ESBLs, class C β-lactamase, Polymerase Chain Reaction, Ps. aeruginosa, pyocyanin pigment, Khartoum-Sudan.
Pseudomonas aeruginosa is one of the leading causes of nosocomial infections worldwide with high mortality rates, particularly among immunocompromised patients1. Ps. aeruginosa infections are difficult to treat, due to its extraordinary antimicrobial resistance to all available classes of antimicrobial agents2, including β-lactams, aminoglycosides and fluoroquinolones3. Resistance to β-lactams occurs by different mechanisms, including blaAmpC overexpression due to genetic mutations, mutant gene acquisition, overproduction of efflux system, or low permeability3. β-lactamases refer to enzymes that hydrolyze the amide bond of the β-lactam ring leading to drug inactivation and therapy failure4. β-lactamases are classified molecularly into four groups: class A (extended spectrum β-lactamases (ESBLs)), class B (metallo-β-lactamases), class C (cephalosporinases), and class D (oxacillinases)5.
ESBLs are enzymes that extend their hydrolyzing ability to hydrolyze broad spectrum cephalosporins6 and they also confer resistance to penicillins and narrow spectrum cephalosporins3. ESBLs are inhibited by β-lactamase inhibitors, such as clavulanic acid7. β-lactamases are transformed to ESBLs usually after point mutations in the β-lactamases gene. These mutations alter the substrate specificity because of changes in the amino acid sequences near the enzyme active site5. ESBLs producing Ps. aeruginosa have been reported worldwide in different countries8–17.
AmpC β-lactamases are class C cephalosporinases that mediate bacterial resistance to cephalosporins and cephamycins. They also exhibit low rates of monobactam, cefepime and carbapenem hydrolysis18 and usually resist the inhibition by clavulanic acid4. Normally, AmpC is a chromosomal β-lactamase gene that is regulated by ampR gene and expressed constantly. Point mutations of ampR gene in Enterobacter cloacae activate AmpC that mediate resistance to β-lactams19. In Ps. aeruginosa over expressed AmpC β-lactamase mediate the resistance to broad spectrum cephalosporins3. AmpC β-lactamase in Ps. aeruginosa has also been reported in different countries around the world20–27.
The exact frequency of β-lactamase producing Ps. aeruginosa in Khartoum State, Sudan is unknown; therefore, the aim of this study was to determine the susceptibility pattern to selected antibiotics and to determine the frequency of β-lactamases producing Ps. areuginosa isolates collected in Khartoum State hospitals.
This is a cross-sectional study conducted between February 2017 and October 2017. Ethical approval for the study was obtained from the ethical committee of the College of Medical Laboratory Science, Sudan University of Science and Technology (SUST) (ethical meeting no, SUST/DSR/1EC/EA2/2017; data, 07th January 2017). Written informed consent from participants was waived by the same ethical committee as the study only used previously collected human bio-specimens with limited participant data.
A total of 121 clinical isolates, which initially identified as Ps. aeruginosa and bacteria other than Ps. aeruginosa was excluded. Selected isolates were obtained from Soba Teaching Hospital, Elribat University Hospital, National Laboratory for Public Health, Ear Nose Throat Hospital, and Military Hospital in Khartoum State. When there was a positive confirmation of Ps. aeruginosa, the study supervisor went and collected the sample from the hospital. The samples were collected from patients suffering from urinary tract infections, respiratory tract infections, blood infections, and wound and ear infections. Data pertaining to the site of infection was collected from hospital records. The bacteria were preserved in 20% glycerol and peptone water and stored at -20°C
Phenotypic identity of the isolates was confirmed through conventional bacterial identification methods, such as Gram stain, oxidase test, and reactions in media containing sugars, such as Kligler Iron Agar, urease test and, citrate test. Pigment production was assessed using Muller Hinton agar, and then phenotypic identity confirmed by genotypic characterization using multiplex PCR, as previously described1.
Muller Hinton medium (HiMedia, India) was prepared and sterilized as instructed by the manufacturer. Antimicrobial susceptibility testing was performed following the modified Kirby-Bauer disc diffusion method1 and the results interpreted according to Clinical Laboratory Standards Institute guidelines (CLSI, 2007). The following antimicrobial discs (HiMedia, India) were used for sensitivity testing: Amoxicillin (25 µg), Cefotaxime (30 µg), Amoxicillin-Clavulanic acid (30 µg), Gentamicin (10 µg), Ciprofloxacin (5 µg), Chloramphenicol (30 µg) and Imipenem (10 µg). Ps. aeruginosa ATCC 27853 was used as quality control strain to control the performance of the test and ensure that the test is properly performed.
Phenotypic identity of the isolates was confirmed through conventional bacterial identification methods such as Gram stain, oxidase test, reactions in media containing sugars such as Kligler Iron Agar, urease test and citrate test.
Genomic DNA was extracted by simple boiling method18. The extracted DNA was used as a template for amplification of target genes using multiplex PCR using TECHNE TC-312 (UK) thermocycler. Firstly, oprI and oprL primers (Table 1) (Macrogen, Korea) were used to confirm the identification of Ps. aeruginosa. Seven primer pairs (Table 1) (Macrogen, Korea) were used for detection of ESBLs genes (blaTEM, blaSHV, blaCTXM-1, blaVEB, blaOXA-1) and class C genes (blaAmpC and blaDHA). The oprI and oprL reaction was carried out with the following cycling conditions: denaturation at 95°C for 5 min; 33 cycles of denaturation at 95°C for 30 secs, annealing at 58°C for 30 sec and extension at 72°C for 30 secs, and final extension at 72°C for 5 min28.
| Target gene | Primers | Sequences (5'-3') | Product size (bp) | References |
|---|---|---|---|---|
| oprI | F | ATGAACAACGTTCTGAAATTC | 250 | 29 |
| R | CTTGCGGCTGGCTTTTTCCAG | |||
| oprL | F | ATGGAAATGCTGAAATTCGGC | 500 | 29 |
| R | CTTCTTCAGCTCGACGCGACG | |||
| TEM | F | ATGAGTATTCAACATTTCCGTG | 861 | 30 |
| R | TTACCAATGCTTAATCAGTGAG | |||
| SHV | F | TTTATGGCGTTACCTTTGACC | 1050 | 30 |
| R | ATTTGTCGCTCTTTACTCGC | |||
| CTXM-1 | F | GACGATGTCACTGGCTGAGC | 499 | 31 |
| R | AGCCGCCGACGCTAATACA | |||
| AmpC | F | ATCAAAACTGGCAGCCG | 550 | 32 |
| R | GAGCCCGTTTTATGGACCCA | |||
| DHA | F | AACTTTCACAGGTGTGCTGGGT | 405 | 33 |
| R | CCGTACGCATACTGGCTTTGC | |||
| VEB | F | CATTCCCGATGCAAAGCGT | 648 | 34 |
| R | CGAAGTTTCTTTGGACTCTG | |||
| OXA-1 | F | GGCACCAGATTCAACTTTCAAG | 564 | 35 |
| R | GACCCCAAGTTTCCTGTAAGTG |
β-lactamases detection was done in two batches; the first batch was used for detection of blaTEM, blaSHV, blaCTXM-1, blaAmpC and blaDHA, while the second batch was used for detection of blaVEB and blaOXA-1 genes. The first batch detection was done in 20 µl of final reaction mixture using Maxime PCR Premix kits (iNtRON Biotechnology, Korea), containing 13 µl of double distilled water (DDW), 0.3µl of each five forward and 0.3 µl of each five reverse primers (1.5 µl), 2 µl of Dimethyl sulfoxide (DMSO) and 2 µl of template DNA.
Amplification of the second batch was done in 20µl using Maxime PCR Premix kits (iNtRON Biotechnology, Korea), containing 14.4 µl of DDW, 0.4 µl of each two forward and 0.4 µl of each two reverse primers (0.8 µl), 2 µl of DMSO and 2 µl of template DNA. Cycling conditions for both amplification reactions were as follows: initial denaturation at 94°C for 2 minutes, then 35 cycles of denaturation at 94°C for 30 secs, annealing at 54°C for 30 secs and extension at 72°C for 50 secs, and final extension at 72° for 5 minutes.
Amplified products were analyzed by electrophoreses at 80 volts for 20 minutes on 1.5% agarose gel containing ethidium bromide and then visualized using UV transilluminator (Uvitec–UK) with 50bp or 100bp molecular DNA ladder (iNtRON Biotechnology, Korea).
From 121 clinical isolates collected from different hospitals in Khartoum State, only 80 (66%) were confirmed as Ps. aeruginosa through conventional methods and species-specific primers; the remaining 41 (34%) isolates were considered as other Gram-negative rod bacteria. The distribution of clinical isolates according to site of infection was as follows: urine, 34 (42%); wound swab, 24 (30%); ear swab, 8 (10%); sputum, 8 (10%); and blood, 6 (7.5%).
The results of antimicrobial susceptibility test for selected antibiotics are presented in Table 2.
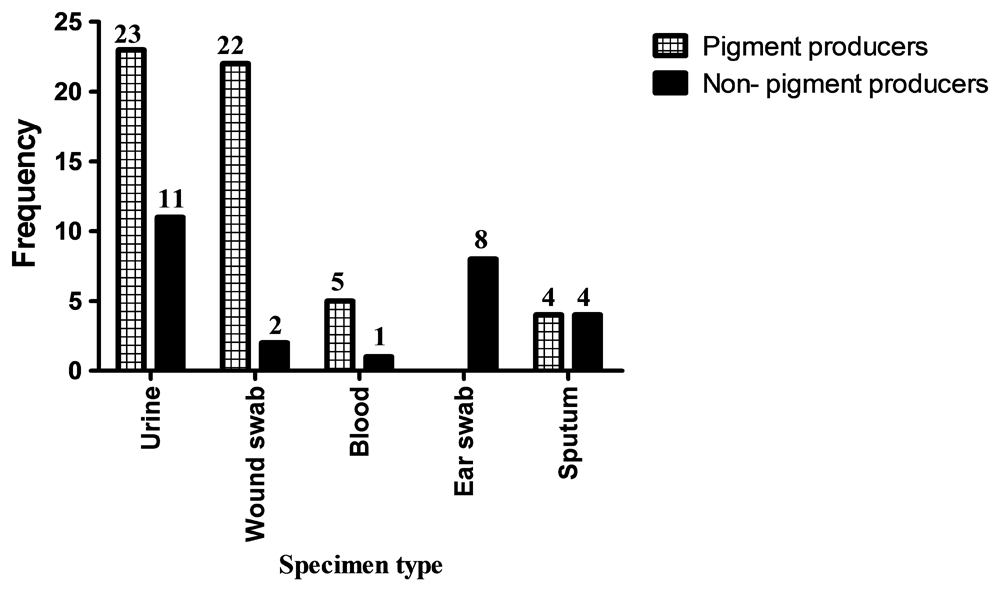
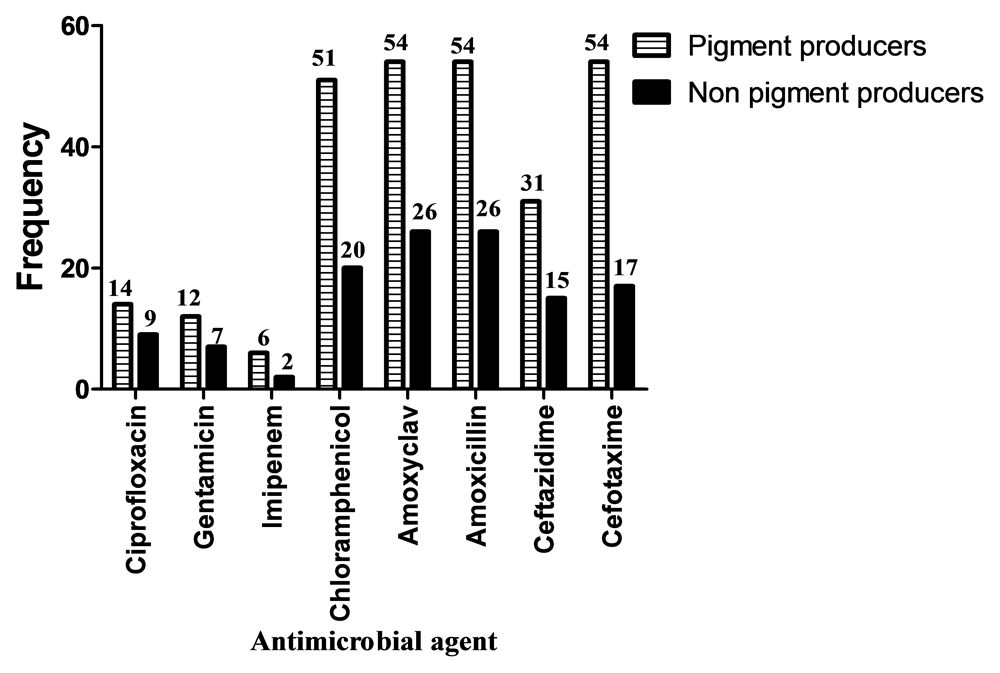
Out of 80 Ps. aeruginosa isolates, 54 (68%) were Pyocyanin pigment producers, while 26 (32%) were not pigment producers. There was a significant association between pyocyanin pigment production and site of infection (P=0.000) (Figure 1). There was also a significant association between pigment production and resistance to Chloramphenicol (P=0.020) and Cefotaxime (P=0.000), while there was insignificant association between pigment production and resistance to other antimicrobials used in this study (Figure 2).
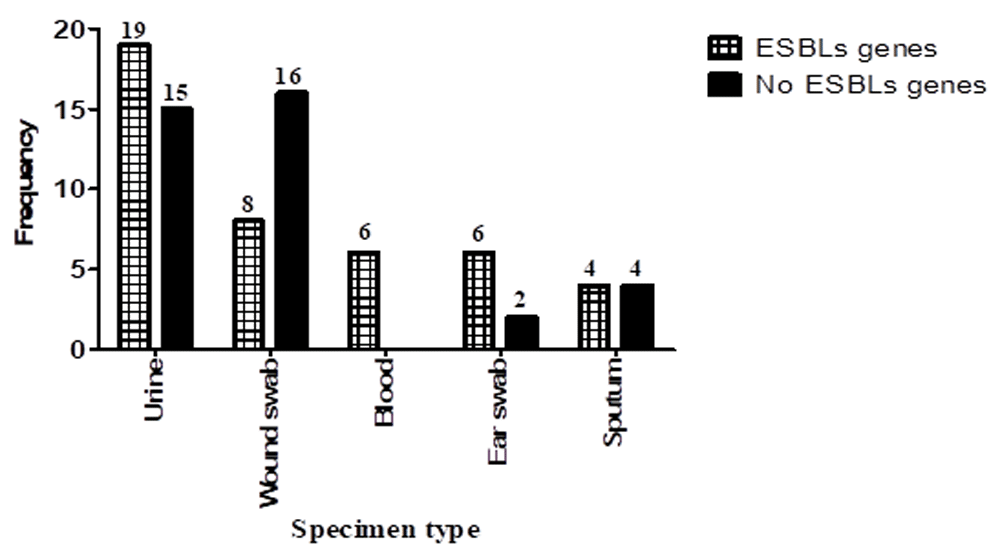
Molecular detection of ESBLs showed that 43 (54%) of the isolates were positive for at least one ESBLs gene, while 37 (46%) were negative for all genes (Figure 3). The frequency of ESBLs gene presence among Ps. aeruginosa was as follows: blaTEM, 19 (44.2%); blaSHV, 16 (37.2%); blaCTX-M1, 10 (23.3%); blaVEB, 14 (32.6%); and blaOXA-1, 7 (16.3%). There was a significant association between the presence of ESBL genes in Ps. aeruginosa and site of infection (P=0.030) (Figure 3). Co-presence of more than one ESBLs gene among Ps. aeruginosa clinical isolates is presented in Table 3.
| blaTEM | blaSHV | blaCTXM-1 | blaVEB | blaOXA-1 | |
|---|---|---|---|---|---|
| blaTEM | 7 | 6 | 5 | 3 | 1 |
| blaSHV | 6 | 4 | 4 | 4 | 1 |
| blaCTXM-1 | 5 | 4 | 0 | 1 | 1 |
| blaVEB | 3 | 4 | 1 | 3 | 3 |
| blaOXA-1 | 1 | 1 | 1 | 3 | 2 |
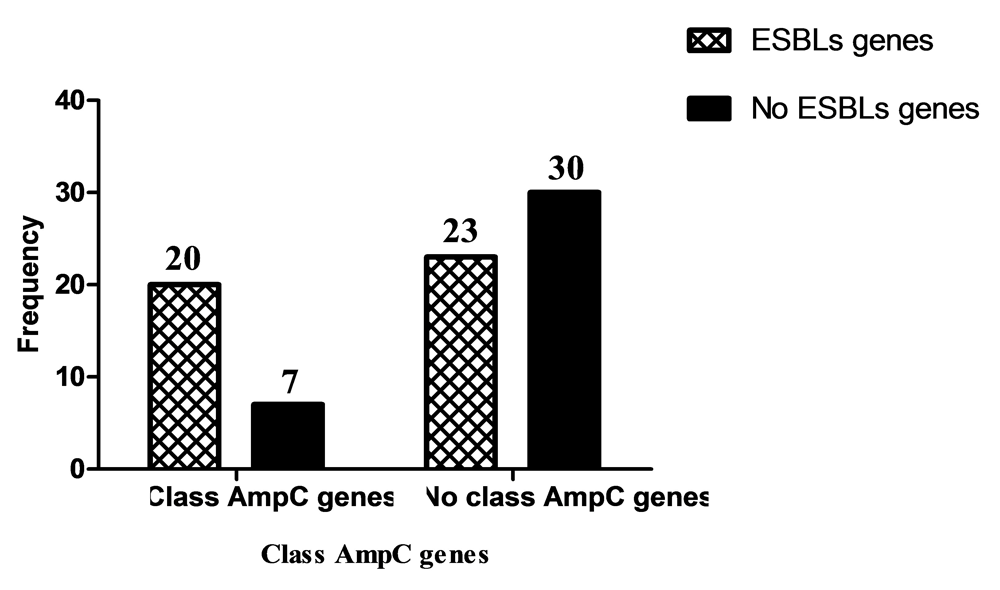
Class C β-lactamases gene were positive in 27 (34%) Ps. aeruginosa isolates; while 53 (66%) were negative (Figure 4). The frequency of class C β-lactamases genes was as follows: blaAmpC, 22 (81.5%); blaDHA, 8 (29.6%); and 3 (11.1%) isolates were positive for both genes. Association between presence of Class C β-lactamases genes and site of infection was insignificant (P=0.215) (Figure 4).
In total, 25% of Ps. aeruginosa isolates were positive for both ESBLs and class C β-lactamase genes (Figure 5). Co-presence of ESBLs genes and class C β-lactamase among Ps. aeruginosa clinical isolates is presented in Table 4.
Pseudomonas aeruginosa is one of the main causative agents of serious nosocomial infections with increased reports of β-lactams resistant strains that makes treatment difficult and complicated19. Production of β-lactamases is one of the most common mechanisms of β-lactam resistance36.
In this study the frequency of ESBL genes among Ps. aeruginosa isolates was 53.8%. This frequency is close to a report from India by Qureshi and Bhatnagar (2016)37, where the frequency of ESBLs in Ps. aeruginosa isolates was 46%. However, the results of this study disagree with another report from India by Gupta et al. (2016)38 where the frequency of ESBLs in Ps. aeruginosa isolates was 22.9%. These discrepancies could be due to differences in strains of the clinical isolates, the antibiotics used or sample size.
The most abundant gene among the ESBLs producing Ps. aeruginosa isolates detected in this study was blaTEM gene (19, 44.2%), followed by blaSHV, blaVEB, blaCTXM-1 and blaOXA-1 (16 (37.2%), 14 (32.6%), 10 (23.3%) and 7 (16.3%), respectively). Similar results were reported by Salah et al. (2016)39 in Egypt concerning the presence of blaTEM, blaSHV and blaOXA-1 (50%, 33% and 17%, respectively). The frequency of blaTEM gene is also similar to that reported by Rafiee et al. (2014)40, which was present in 39.2% of isolates. In a study in Iran by Sales et al. (2017)41, similar results were reported concerning the presence of blaCTXM-1 (27.3%), while in India Jamali et al. (2017)15 reported a higher frequency (57.5%) of this gene.
On the other hand, the frequency of blaVEB and blaSHV genes in this study differs from that reported in Iran by Bokaeian et al. (2014)42 where blaVEB gene frequency was 13.3%, while the frequency of blaSHV gene was 6.6%. The report by Jamali et al. (2017)15 concerning the genes blaTEM (15%) and blaSHV (75%) are also different from those reported in this study, and this could be due to the variation in strains of clinical isolates and the sample size used.
In this study, the frequency of class C β-lactamase genes in Ps. aeruginosa isolates was 27 (34%). This result is close to a report from India by Gupta et al. (2016)38 where 43% of Ps. aeruginosa were AmpC producers, and disagrees with a report from Thailand by Katvoravutthichai et al. (2016)43 where 11% of Ps. aeruginosa isolates were AmpC producers. In this study, out of the class C β- lactamase producing Ps. aeruginosa isolates, 22 (81.5%) and 8 (29.6%) isolates were positive for blaAmpC and blaDHA genes, respectively. Qureshi and Bhatnagar (2016)37 in India reported that no Ps. aeruginosa isolates were positive for blaAmpC gene, while Rafiee et al. (2014)40 in Iran reported that 60.8% of Ps. aeruginosa were positive for blaAmpC gene.
All Ps. aeruginosa clinical isolates tested in our study were resistant to Amoxicillin and Amoxyclav and this may be due to the misuse of antibiotics in Sudan44, where plenty of antimicrobial agents are sold over the counter. This rate of resistance is higher than the rate of resistance reported by Ahmad et al. (2016)45 in Pakistan where the resistance to Amoxicillin and Amoxyclav was 73.4% and 67.7% respectively, and this could be justified by the time difference between the studies, as well as the difference in the strains and antibiotics used.
The resistance rate of Ps. aeruginosa to Imipenem was 10% (n=8). This may be due to the infrequent use of Imipenem antibiotics. This percentage agrees with a study reported in Pakistan by Ahmad et al. (2016)45 where 11.1% of Ps. aeruginosa isolates were resistant to Imipenem, while in Sudan Altom and Ahmed (2015)46 reported that 5.7% Ps. aeruginosa isolates were resistant to Imipenem. This finding may indicate that carbapenem resistance is on the rise in Ps. aeruginosa isolates from Sudan.
In this study, the number of Ps. aeruginosa isolates resistant to Cefotaxime, Chloramphenicol and Ceftazidime was 71 (88.8%), 71 (88.8%) and 46 (57.5%), respectively. The rate of resistance to Cefotaxime in this study is different from that reported in Pakistan by Ahmad et al. (2016)45 who found that 20.3% of Ps. aeruginosa isolates were resistant to Ceftazidime. The resistance rate in this study also disagrees with that reported by Albadawi (2010)47 in Sudan who found that resistance of Ps. aeruginosa to Ceftazidime and Cefotaxime were 31% and 42%, respectively. These findings also indicate the rapidly increasing rates of Ps. aeruginosa resistance to antimicrobial agents in Sudan perhaps due to antibiotic misuse.
In this study, 23 (28.8%) and 19 (23.8%) of Ps. aeruginosa clinical isolates were resistant to Ciprofloxacin and Gentamicin, respectively. Altom and Ahmed (2015)46 in Sudan also reported that 18.6% of Ps. aeruginosa were resistant to Gentamicin. Different results were reported by Ahmad et al. (2016)45, where the percentage of resistance to Gentamicin and Ciprofloxacin were 74.3% and 44%, respectively. These percentages are much higher than those reported in this study probably due to their different geographical location, study time difference and the antibiotic-use rates.
In this study, there was significant association between ESBLs production and site of infection (P=0.030). Ps. aeruginosa isolated from blood showed the highest ESBLs production followed by ear swab, urine, sputum and wound swab. This result agrees with a study in India reported by Basak et al. (2012)48 where the highest ESBLs producing Ps. aeruginosa isolates were from blood, but disagrees with Azizi et al. (2015)49 in Iran who found that highest ESBLs production was in Ps. aeruginosa isolated from wound followed by urine, sputum and blood. There was insignificant association between the presence of class C β-lactamase genes and site of infection (P=0.215) found in the present study. The highest frequency of class C β-lactamases genes in Ps. aeruginosa were isolated from blood followed by urine, ear swab, wound swab and sputum. These results agree with the study in India reported by Basak et al. (2012)48 in that the highest percentage of class C β-lactamase genes in Ps. aeruginosa were found in blood.
In the present study, there were four clinical isolates phenotypically sensitive to third generation cephalosporins (Ceftazidime and Cefotaxime) and genotypically positive for ESBLs genes. This result indicates that Ps. aeruginosa may carry hidden unexpressed genes that could be detected through molecular techniques. This result agrees with a study in India reported by Bajpai et al. (2017)50 where out of 38 phenotypically ESBL-negative isolates, 20 isolates were positive for ESBLs genes.
In this study, the frequency of pigment producing Ps. aeruginosa isolates was 67.5%. In a study in India a higher percentage was reported where 82.5% of Ps. aeruginosa were pigment producers (Finlayson and Brown, 2011)51. The present study revealed that there was no relationship between pigment production and pattern of antimicrobial resistance, except in Chloramphenicol (P=0.02) and Cefotaxime (P=0.00).
In this study, the relationship between pigment production and site of infection was significant (P=0.000). Ps. aeruginosa isolated from wound infections were the highest pigment producing isolates followed by isolates from blood, urine and sputum. There was no pigment production in Ps. aeruginosa isolated from ear infections in this study.
The variations between the results of this study and other reports could be attributed to the difference in antibiotic usage patterns in each region, economical causes, geographical differences, sample size, differences in time in which the studies were performed, and study population. Despite the significance of the present study, there were limitations that should be avoided in future studies, such as the small sample size, phenotypic detection of β-lactamases, coverage of other β-lactamase classes, and gene sequencing should be done in order to confirm and to identify all the genes that are carried by Ps. aeruginosa strains in Sudan.
This is study is of great importance as it raises attention to the existing problem of resistance to β-lactams in Ps. aeruginosa in Sudan. This study confirms the reports that a number of antibiotics are becoming useless for treating this problematic bacterium, since all the strains of Ps. aeruginosa isolates in this study were resistant to Amoxicillin and Amoxyclav. The best antibiotic sensitivity results obtained in this study were those of Imipenem, followed by Gentamicin and Ciprofloxacin. Moreover, Ps. aeruginosa isolates showed an increased rate of β-lactamase production with co-resistance with other classes of antibiotics. Of interest is the finding that clinical isolates were resistant phenotypically in high frequencies to Amoxicillin, Amoxyclav and a third generation antibiotic, Cephalosporin, and showed negative results genotypically, indicating that resistance to this family of antibiotics also exist by resistance mechanisms other than β-lactamases production. Also, our PCR results revealed that Ps. aeruginosa possesses hidden β-lactamases genes that can’t be detected phenotypically. Finally, this study highlighted for the first time the problem of misidentification of Ps. aeruginosa and other microorganisms in Khartoum hospitals as only 80 out of the 120 alleged isolates were confirmed to be Ps. aeruginosa through PCR.
Figshare: SPSS, https://doi.org/10.6084/m9.figshare.12453287.v251
This project contains the following underlying data:
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
Deep thanks to all the Microbiology Department team at Sudan University of Science and Technology for their significant help during the study. We are especially thankful to Mariam Awad Ahmed Suliman for her kind support and great advice throughout the study and very special thanks to Dr. Ahmed Bakheet Abd Alla for his kind guidance and support.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: microbiology, public health
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 3 (revision) 01 Apr 21 |
||
|
Version 2 (revision) 15 Sep 20 |
read | read |
|
Version 1 27 Jul 20 |
read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)