Keywords
Serine proteases, human proteinase 3, house dust mites group 3 allergens, ANCA associated vasculitis, sequence homology, T and B cell epitopes, cross-reactivity, epitope modelling
This article is included in the Cell & Molecular Biology gateway.
This article is included in the Bioinformatics gateway.
Serine proteases, human proteinase 3, house dust mites group 3 allergens, ANCA associated vasculitis, sequence homology, T and B cell epitopes, cross-reactivity, epitope modelling
The manuscript was revised and refined by a language style corrector. Some sentences in the methods and results section were improved giving more detail and explanations in accord to revisor suggestions.
See the authors' detailed response to the review by Chandrabose Selvaraj
Anti-neutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV) is a life-threatening autoimmune disease affecting small vessels, compromising the respiratory mucosa, skin, lung, and the kidney1. This group of small vessel vasculitis includes various diseases: granulomatosis with polyangiitis, microscopic polyangiitis, kidney-limited vasculitis and Eosinophilic granulomatosis with polyangiitis, all of them having in common some degree of autoimmune response to the Human Proteinase 3 protein (PR3). Previous studies have shown that autoantibody binding to PR3 expressed on the neutrophil surface may activate its degranulation, eliciting tissue damage in small vessels and their irrigated organs. Also, while proinflammatory effector T cells have been implicated in vasculitis pathogenesis2, a specific PR3 T cell epitope has not been reported in AAV patients3. PR3 is a serine protease physiologically expressed in human neutrophils. Due to its enzymatic activity, it degrades various intercellular gap-junction proteins and collagen and may play a role in neutrophil transendothelial migration. In addition, this protein is an important autoantigen in AAV, and sera from patients with severe and relapsing forms of the disease can bind it in IgG ELISA assays4–6. Further, although a cause-effect relationship between PR3-autoantibodies and vasculitis is not clearly defined, animal models support a pathogenic role7,8, revealing that they may be involved in disease inception, progression and severity1.
Environmental exposures, specially to microbial components mimicking self-antigens have been proposed as triggers of autoimmunity9,10. Also, in AAV, it has been proposed that an endogenous immune response to a complementary protein to PR3 autoantigen could be implicated in disease inception, and this antisense protein harbors homology to various bacterial peptides11. PR3 crystal structure has been elucidated, and various epitopes are recognized by patients suffering AAV; however, its cross-reactivity with environmental antigens is poorly studied12–14.
Previous studies have shown that specific IgE to some self-proteins have been identified in autoimmune and allergic diseases like lupus, urticaria, dermatitis, allergic pulmonary aspergillosis and have a strong association with disease activity15–18. Some allergens can cross-react with human proteins and participates in autoimmunity inception in pemphigus vulgaris by a “hit-and-run” mechanism, opening the theoretical possibility for a similar mechanism to occur in another autoimmune disease such as AAV19–22.
In the tropics, house dust mites (HDM) are important ubiquitous allergen sources and exposure is perennial, increasing the possibilities of exposure in the general population23, and IgE sensitization to their components24,25. Sensitization to HDM group 3 allergens is common26, as they harbor serine protease activity and conserved structural homology27, making them potential PR3 cross reactive antigens; this has not been explored before. Here, we show in silico data suggesting cross-reactivity and epitope sharing between PR3 and HDM group 3 allergens.
The amino acid sequence from the human PR3 (Uniprot accession: P24158) was used as query to perform a search for serine protease homologous reported in allergenic sources: Dermatophagoides pteronyssinus (Der p 3: Accession number P39675), Blomia tropicalis (Blo t 3: A1KXI1), Glycyphagus domesticus (Gly d 3: Q1M2M8), Lepidoglyphus destructor (Led p 3: Q1M2L7) and Tyrophagus putrescentiae (Tyr p 3: C6ZDB5) with the PSI-BLAST tool. Parameters were set as default.
Identity among all allergenic sequences homologous to PR3 was analyzed using the Jalview tool2.11.028. First, all allergens and human PR3 codes were used as inputs in the Jalview tool. Second, the T coffee tool was chosen to assess alignment. Third, alignment was displayed as an identity percentage.
The 3D model of Der p 3, a serine protease of Dermatophagoides pteronyssinus was generated by homology in the SWISS-MODEL server using the zymogen catalytic region of human MASP-2 (PDB: 1zjk f) as a template. The 3D model of Der p 3 was loaded into the ProSA-web server29, which was used to analyze its quality.
The model was refined in DeepView v4.1 (energy minimization and rotamer replacements). Its quality was evaluated by several tools, including Ramachandran graphs, WHATIF, QMEAN4 index, and energy values (GROMOS96 force field). For the validation of the Der p 3 structure we used the Minimize Structure option in the UCSF Chimera software, a procedure that adjust the energy and reduce the entropy of the model30.
Three-dimensional structure (PDB: 1FUJ) of the human PR3 serine protease was retrieved from the Protein Data Bank. A cartoon model was created using Pymol software v2.4. Root median square deviation (RMSD) value between Der p 3 and PR3 was calculated using Chimera software v1.030.
ElliPro v3.0 and BepiPred v2.0 tools were used to predict B and T cell epitopes on Der p 331,32. With ElliPro, the 3D structure of Der p 3 was used to predict epitopes. Minimum score and maximum distance (Angstrom) were set to 0.5 and 6, respectively. Epitopes with high conserved rates were visualized in the 3D model. For prediction using BepiPred, an amino acid sequence of Der p 3 was used as input.
The 3D structure of Der p 3 was submitted to the ConSurf server to generate evolutionarily related conservation scores to help to identify functional regions in the proteins. HMMER algorithm, 1 iteration, E-value cutoff (0.0001) and UNIREF-90 database was set as default to generate multiple alignment, prior to evolutive analysis. All amino acid sequences in FASTA format were used.
BLAST search identified various serine protease family members from HDM as homologous. The multiple sequence alignment analysis showed that Der p 3, Blo t 3, Gly d 3, Led p 3 and Tyr p 3 allergens shared 45% of identity in their aminoacid sequences with PR3. The most conserved region is located between residues 53 to 75, indicating the existence of molecular mimicry (Figure 1). Among the members of HDM group 3 allergens, an identity until 41% was reported (Table 1), and a highly conserved region between residues 40 to 90 was found. When identity between PR3 and each allergen used in study was analyzed, a moderate level of identity was found (30%) (Table 1).

An identity of 45% in their amino acid sequences was found.
All comparisons of PR3 with HDM group 3 allergens showed a moderate identity.
| PR3 | Der p 3 | Blo t 3 | Gly d 3 | Lep d 3 | Tyr p 3 | |
| PR3 | 100 | 33 | 27 | 30 | 30 | 27 |
| Der p 3 | 33 | 100 | 48 | 52 | 53 | 43 |
| Blo t 3 | 27 | 48 | 100 | 58 | 58 | 47 |
| Gly d 3 | 30 | 52 | 58 | 100 | 99 | 41 |
| Lep d 3 | 30 | 53 | 58 | 99 | 100 | 41 |
| Tyr p 3 | 27 | 43 | 47 | 41 | 41 | 100 |
A structural model of Der p 3 was obtained by homology modelling using the 3D structure of PR3 reported in the PDB database. According to modelling, the Der p 3 tertiary structure exhibited a typical fold of serine protease family, conformed by four α-helixes and fifteen β-strands with structural homology with PR3 (RMSD = 0.8) (Figure 2).
Using ElliPro and BepiPred servers, a cross reactive B cell epitope was predicted on all serine protease used in this study. This epitope is formed by ten residues and is on the N-terminal region, spanning amino acids 29 and 39 with a surface area of 470 Å, not forming part of any domain within the protein. Conservative analysis indicated that the antigenic region predicted was highly conserved in the serine proteases (Figure 3). According to ConSurf analysis, the region covering the cross-reactive epitope is conserved among the serine protease family (Figure 4). T cell epitope prediction identified at least two epitopes with potential cross-reactivity among all sequences analyzed. Both epitopes are located on the first and second β strands: the first epitope spans the 45 to 59 region (ISLQSSSHFCGGTIL); and the second, the 63 to 77 region (WILTAAHCVAGQTAS) (Figure 5; Table 2).

(A) Surface model of Der p 3, showing area occupied by B cell epitope predicted as cross reactive. (B) Cartoon model showing location of epitope on tridimensional structure. It can be appreciated that the predicted epitope is on a loop spanning residues 29 to 39.
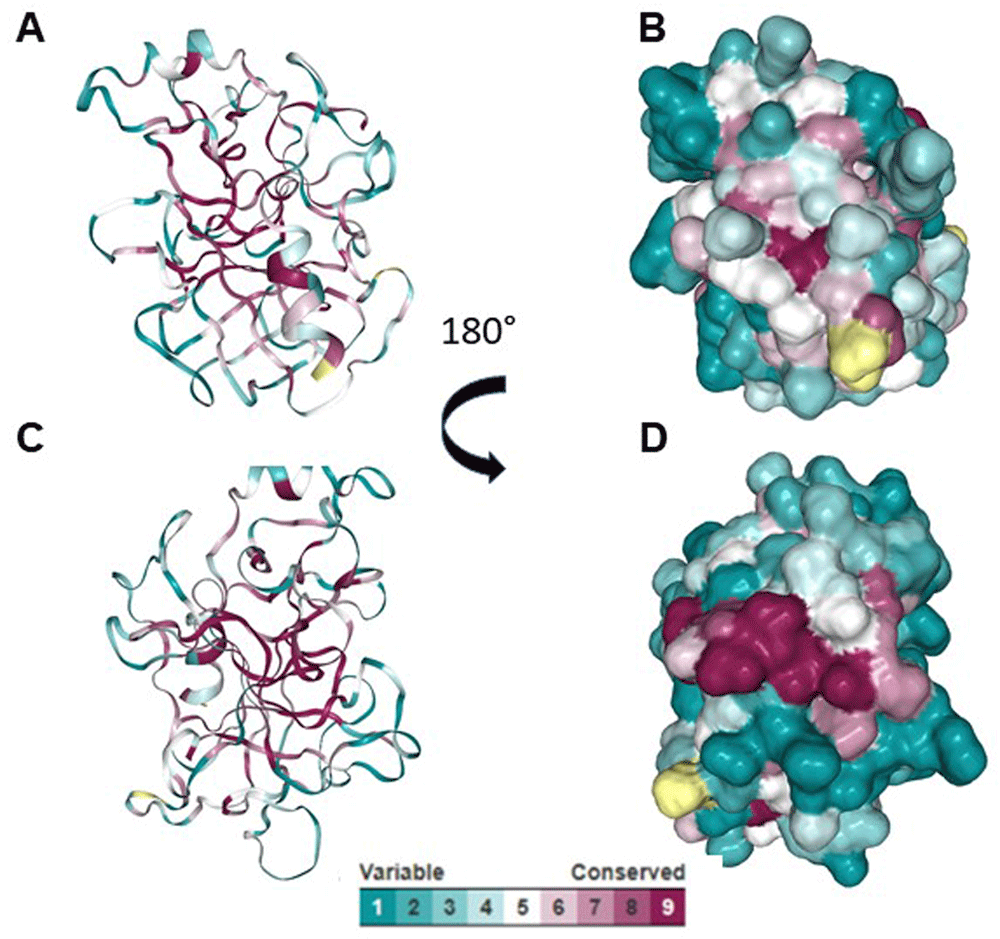
(A and C) Cartoon models showing the conserved region among serine proteases. (B and D) Surface models showing the conserved region among serine proteases.
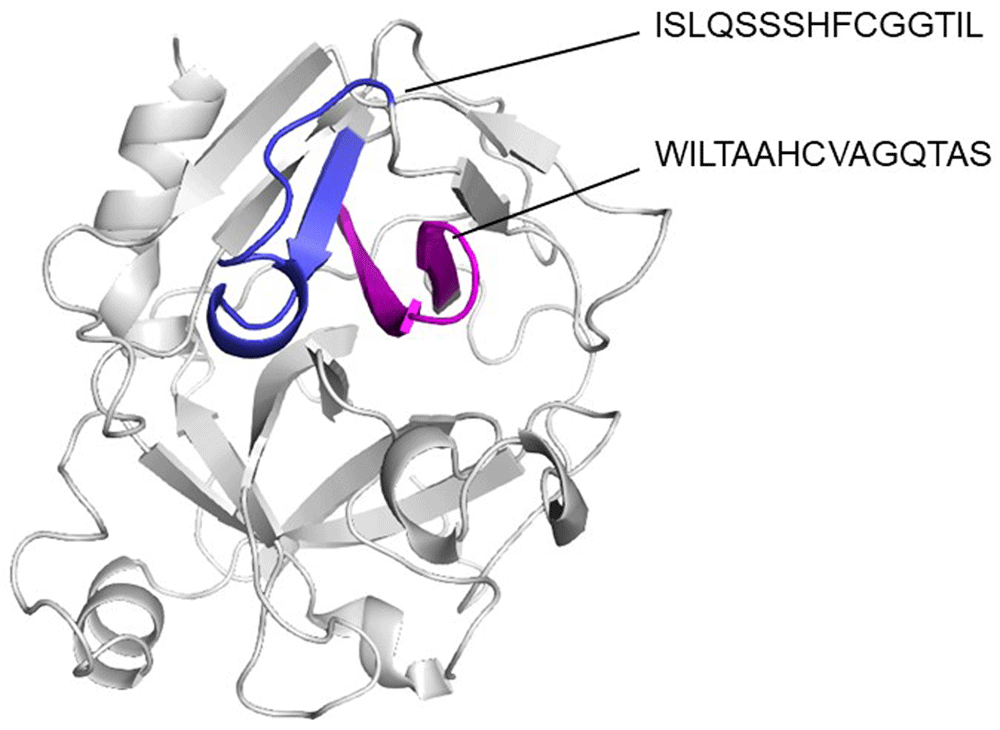
It can be appreciated that predicted epitopes are in a continuous β strands (blue and magenta).
In this study we found that PR3 and HDM group 3 serine protease allergens have conserved identity and homology. Also, for the first time, we predicted various T and B cell cross reactive epitopes between them through an in silico approach. PR3 is an important autoantigen in small vessel vasculitis and it seems to participate in disease inception, progression, and severity1. Our results have potential implications for the understanding of autoreactive response in AAV and open the possibility for a new environmental trigger of the autoreactive response in AAV.
In AAV, it has been proposed that autoantibodies directed to a complementary protein to PR3 autoantigen could be implicated in disease inception, and this antisense protein harbors homology to various bacterial peptides11 – a theory named autoantigen complementarity33. However, in epidemiological studies, autoantigen complementarity hypothesis testing has showed conflicting results, since sera from some patients suffering from AAV do not recognize complementary PR3, while others do34–36. Also, molecular mimicry of PR3 protein by infectious microorganism components have been proposed as a possible environmental trigger of the disease based on the reports of infections preceding the manifestations of vasculitis10,37–39, although a cross reactive antigen have not been reported yet.
In their seminal publication, Pendergraft et al. run a BLAST query to find homologues of PR3 protein in microbial or fungal microorganisms, and do not find matching sequences at that time11. However, they do not include Arachnida or other environmental sources of cross-reactivity. In our analysis we find matching PR3 protein sequences with various HDM group 3 serine protease allergens, and at least theoretically this finding could have many implications for the understanding of inception and even diagnosis of autoreactive response in AAV. Recently, Qian et al. have shown that some allergens can cross-react with human proteins19 and participate in autoimmunity inception in pemphigus vulgaris by a “hit-and-run” mechanism22, opening the theoretical possibility for a similar mechanism to occur in another autoimmune disease such as AAV. Similarly, in atopic dermatitis, Valenta and collaborators observe that some patients with severe complications from the disease, had IgE directed to the profilin of the Betula verrucosa, but also to the human homologue40.
In the tropics, HDM are important ubiquitous sources of protease allergens. Exposure is perennial, increasing the possibilities of exposure and IgE sensitization to their components in the general population23–25. Sensitization to HDM group 3 allergens is common26, and they harbor serine protease activity27, a characteristic that make them highly allergenic. Moreover, their conserved structural homology makes them highly immunogenic41,42 and suitable for epitope spreading43. In this context, “hit-and-run” and epitope spreading establish framework mechanisms for environmental allergens with homology to autoantigens to potentially participate in the development of autoimmunity. We speculate that HDM group 3 allergens harbor two characteristics that make them suitable candidates for environmental triggering of AAV: their proteolytic activity that, as other protease allergens, set a tissue damaging microenvironment during antigen recognition41; and molecular homology-epitope sharing with human PR3, that would elicit B cell autoantibody production and autoreactive T cell receptor generation. In conclusion, we observe that PR3 and HDM group 3 serine protease allergens have conserved identity, and for the first time we predict cross-reactive epitopes between them through an in silico approach.
UniProtKB: PRTN3_HUMAN, Accession number P24158: https://www.uniprot.org/uniprot/P24158
Protein Data Bank: PR3 (MYELOBLASTIN), Accession number 1FUJ: https://www.rcsb.org/structure/1FUJ
UniProtKB: Mite allergen Der p 3, Accession number P39675: https://www.uniprot.org/uniprot/P39675
UniProtKB: Trypsin Blo t 3, Accession number A1KXI1: https://www.uniprot.org/uniprot/A1KXI1
UniProtKB: Gly d 3, Accession number Q1M2M8: https://www.uniprot.org/uniprot/Q1M2M8
UniProtKB: Allergen Lep d 3, Accession number Q1M2L7: https://www.uniprot.org/uniprot/Q1M2L7
UniProtKB: Trypsin Tyr p 3.0101, Accession number C6ZDB5: https://www.uniprot.org/uniprot/C6ZDB5
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Nephrology, Rheumatology, Clinical immunology, Pediatrics
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Erban T, Klimov P, Harant K, Talacko P, et al.: Label-free proteomic analysis reveals differentially expressed Wolbachia proteins in Tyrophagus putrescentiae: Mite allergens and markers reflecting population-related proteome differences. Journal of Proteomics. 2021; 249. Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Invertebrate physiology, proteomics, acarology, allergens, multiomics
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
No
If applicable, is the statistical analysis and its interpretation appropriate?
No
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Immunoinformatics, Computational Biology, Molecular Modeling
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 03 Feb 22 |
read | read | |
|
Version 1 26 Jan 21 |
read | ||
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)