Keywords
Plasmodium falciparum, Plasmodium vivax, malaria, animals, host reservoir, PCR.
Plasmodium falciparum, Plasmodium vivax, malaria, animals, host reservoir, PCR.
Malaria is transmitted by the Plasmodium vector Anopheles mosquitoes. Four Plasmodium types, namely P. falciparum, P. vivax, P. ovale, and P. malariae cause pathologic conditions in humans. Recently in Southeast Asia, P. knowlesi infection cases have also been reported.1–3
Before molecular diagnostics development, only humans were assumed to be the primary host for Plasmodium. However, studies in the last two decades on Plasmodium reported that the parasites originated from animals. Further stating that P. falciparum originated in the gorilla4 and chimpanzee,5,6 P. vivax was from African apes,7 P. malariae was from chimpanzees6 and P. knowlesi was from monkeys,8,9 while P. ovale in humans and chimpanzees are genetically identical.10 The factors hypothesized to explain this situation include primate’s habitat loss and human’s aggressiveness in exploring forest.11 A study from South Kalimantan reported the contribution of forest workers to malaria incidence.12
East Nusa Tenggara and West Papua are known as malaria-endemic areas in Indonesia as their annual parasite incidence (API) in 2015 was 31.29% and 7.04%, respectively,13 while in 2018, according to the health office in both the districts, the API rate in Fakfak, West Papua and East Nusa Tenggara, West Sumba was 4.85% and 12.9%, respectively.(unpublished data) Due to this situation, we aimed to explore the presence of human Plasmodium among domestic animals that are a potential reservoir host.
This study was conducted in October 2018 in Gaura village, West Sumba Regency, an area 29.96 km2 in size inhabited by 9,584 people, and Fakfak, West Papua Province, in August 2019 with an area of 11,036 km2 inhabited by 84,692 people (Figure 1). The residents’ main occupation is farming, while livestock such as goats, horses, cows, pigs, and buffalos are commonly found in their enclosures located around the owner’s residence. Furthermore, they also own pets such as dogs and cats.
Sampling was carried out by the veterinarian and staff from West Sumba and Fakfak Animal Husbandry Office. The buffaloes, goats, pigs, and horses’ blood samples were collected in 5 ml EDTA tubes from the jugular vein located in the ventrolateral area of the neck using vacutainer needles, size 16–18. Meanwhile, the dog’s blood was drawn from the cephalic antebrachial vein in the leg using a size 21 vacutainer needle. By using a micropipette, approximately 10 ul of EDTA blood was dropped onto a microscope slide, then smeared and stained with Giemsa (MERCK Millipore, Germany) for Plasmodium microscopic identification, while the remaining was dropped onto a filter paper (Whatman CAT No. 1442-090) until it absorbed to about 1.5 cm in diameter. The dry filter paper was put on a sterile plastic clip and stored at room temperature for a maximum of 10 days.
A dried blood spot (DBS) isolation kit for DNA extraction on filter paper (Cat. no. 36000) from Norgen Biotec was used. A 6 x 3 mm piece of blood-stained filter paper was put into a 1.5 ml tube containing 100 μl of digestion buffer B. It was vortexed and incubated at 85°C. Afterwards, 20 μl of proteinase K and 300 μl of lysis buffer B were added to the tube and then vortexed before incubation at 56°C for 10 minutes. About 250 μl of 95% ethanol was added to the tube and then vortexed, while the DNA content was washed by adding 500 μl of WN wash solution and centrifugated for one minute at 8,000 rpm. Washing was carried out again using 500 μl of WN wash solution and centrifugated at 14,000 rpm. For DNA elution, 90 μl of elution buffer B was put into the tube and centrifuged at 8,000 rpm for one minute, and the purified DNA was stored at -20°C.
DNA amplification of nested PCR and qPCR were performed as directed by Tiangen Biotech (Beijing). Plasmodium DNA amplification was carried out using the nested PCR method with a 2× Tag Plus PCR mix enzyme (Tiangen). The final volume of 12.5 μl contained 6.25 μl enzyme, 2.25 μl ddH2O, 1 μl forward primers, 1 μl reverse primers, and 2 μl DNA sample. For sequencing, the PCR mixture’s volume was doubled, with the final volume being 25 μl, while the primer sequences of P. falciparum, P. vivax14 and P. knowlesi15 can be seen in Table 1.
The nested one DNA amplification temperature was set at 94°C denaturation (one minute), 55°C annealing (one minute) and 72°C extension (one minute) for 35 cycles. For nested two, denaturation was carried out at 94°C (30 seconds) and extension was at 72°C (30 seconds) in 35 cycles. There was a difference in the annealing temperature for each species in nested two, namely 55°C (one minute) for PCR multiplex P. falciparum and P. vivax, but 56°C (one minute) for P. knowlesi. Nested one products were used as templates for nested two and both were run on agarose gel 1.5% and 2%, respectively, while qPCR was analysed using agarose gel 1.5% for electrophoresis. Molecular work was not performed for P. ovale and P. malariae due to difficulties in finding the positive control, and according to the local health office these species have never been reported from Sumba and Fakfak.
Considering the possibility of contamination, DNA was re-extracted from blood from the same filter paper. PCR was performed using the primers, rPF1 and rPF2, as well as rPV1 and rPV216 to detect P. falciparum and P. vivax, respectively. The same extraction and amplification method were used as described above.
To determine the Plasmodium species, in the second round of nested PCR, products having positive band targets were sent to the 1st BASE, Axil Scientific Pte Ltd Singapore for sequencing. The DNA sequence result was adjusted using multiple alignments found in the BioEdit 7.0 application17 and then read by the BLAST program from the NCBI website.
This study was approved for ethical clearance by the ethics committee of the Faculty of Medicine, Hasanuddin University (734/H4.8.4.5.31/PP36-KOMETIK/2018). All efforts were made to ameliorate any suffering of animals. To prevent stress, animals were comforted by their owners while blood samples were taken, and sampling was performed by experienced officers. Second and third blood samples were taken if there was a failure in the first sample and only if the animals were cooperative. About 20% of animals were sampled more than once.
A total of 208 and 62 animal blood samples were collected from Gaura and Fakfak villages, respectively. These consisted of 92 buffalos, 21 horses, 121 goats, 18 dogs, and 18 pigs. Using the nested PCR method, 32 of the 270 animals were found to be P. falciparum and P. vivax positive. The percentage of Plasmodium positive animals included 20.7% buffalo, 14.3% horse, 5.8% goat, and 16.7% dog with one buffalo having a mixed infection (P. falciparum and P. vivax). There was no P. knowlesi found in any of the samples and no other Plasmodium was found in 18 pig blood samples. PCR gel products, DNA sequence results, and the sample’s quality can be seen in Figures 2, 3 and 4, respectively.18 Plasmodium distribution in the animals’ blood samples from Gaura and Fakfak are presented in Table 2, and it shows that blood containing Plasmodium was only found in Gaura. The results of the qPCR using rPF1–rPF2 and rPV1–rPV2 primers were similar to the nested PCR
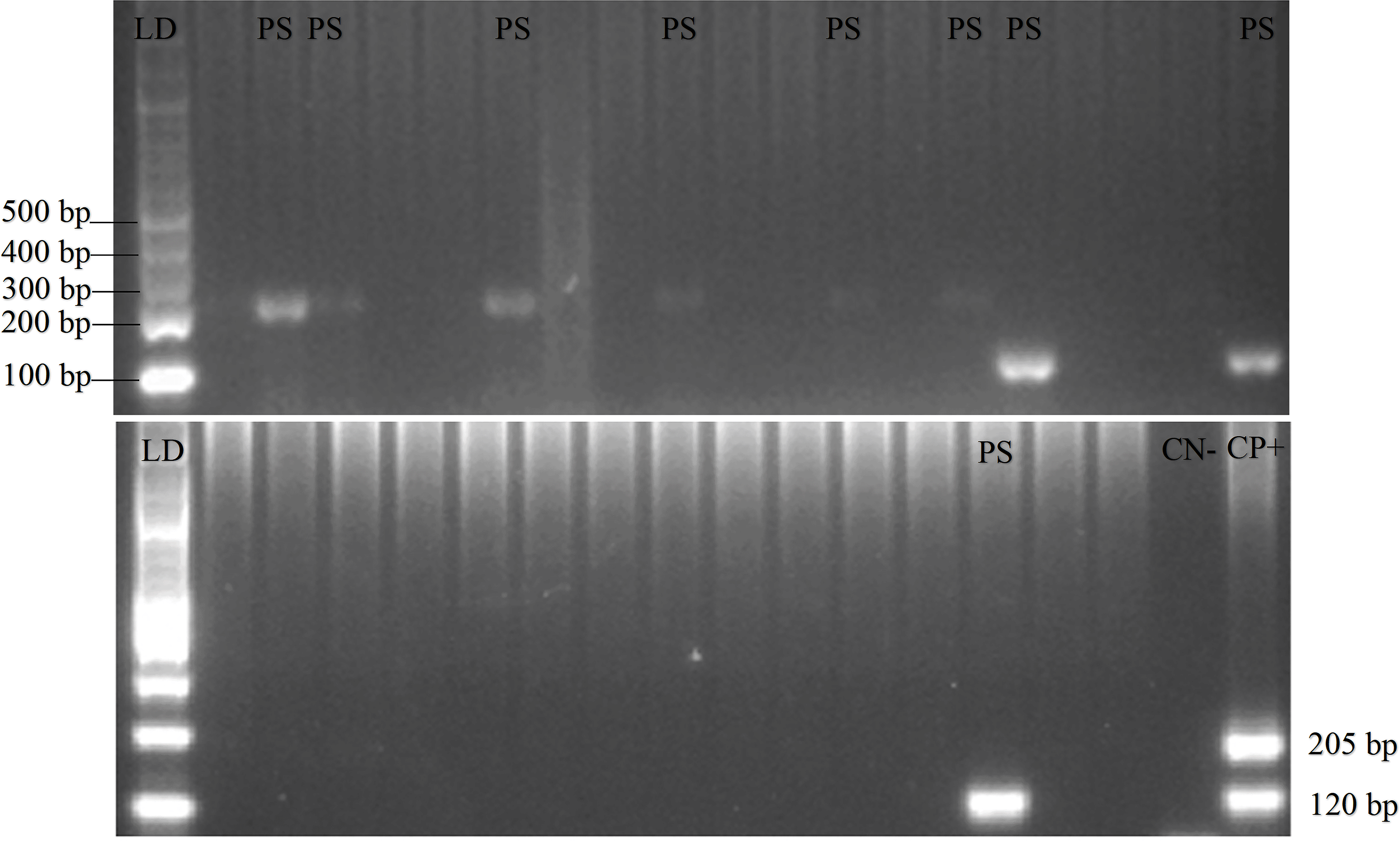
Gel view of PCR product from Plasmodium vivax and Plasmodium falciparum in domestic animals in Gaura, West Sumba (LD = DNA ladder, PS = positive samples, CN = control negative, CP = control positive) by nested PCR (multiplex PCR). 120 bp for positive Plasmodium vivax, 205 bp for positive Plasmodium falciparum.
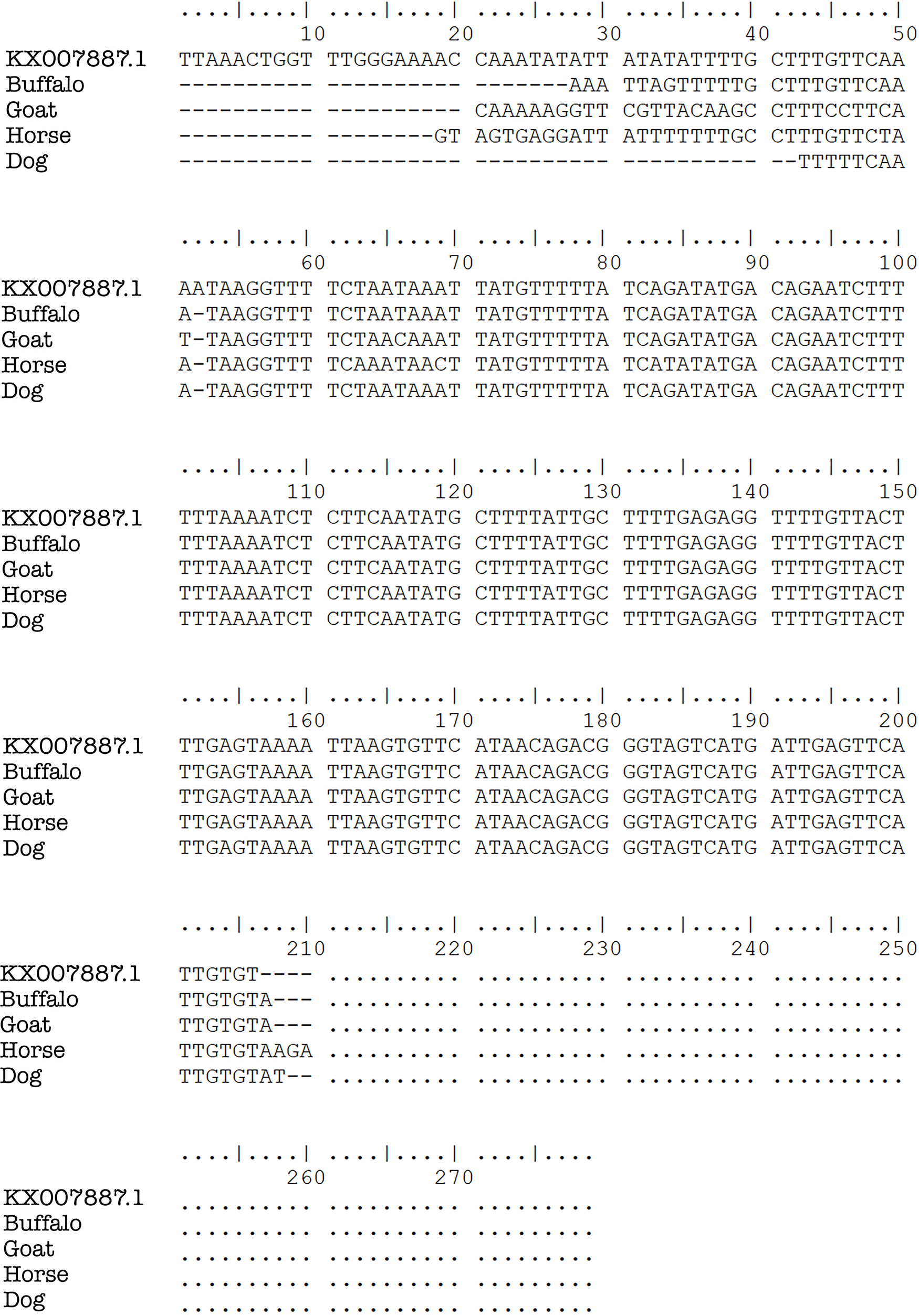
Plasmodium DNA sequence alignments from blood samples taken in Gaura village, West Sumba, Indonesia by ClustalW multiple sequence alignment.
Example of Plasmodium PCR product quality from a blood sample taken in Gaura village, West Sumba, Indonesia.
Distribution of animal blood samples and Plasmodium species found in Gaura village, West Sumba, Indonesia and Fakfak, West Papua, Indonesia.
Microscopically, trophozoites, schizonts, and gametocyte forms at 100× magnification can be seen in Figure 5. P. falciparum gametocytes found in buffaloes were sausage and crescent-shaped (a, b), while schizonts found in horses were smaller or the same size as the red blood cells (c). The P. vivax gametocyte was larger than the red blood cells found in buffalo (d). P. falciparum gametocyte and trophozoites (ring-shaped) with one or two nuclei was found in goats (e) and P. falciparum trophozoite found in horses had one nucleus (f).
The presence of Plasmodium was suspected in domestic animals because malaria cases in these two villages remained high despite maximum preventive efforts having been applied including insecticide-treated bed nets. About 32 of the 270 blood (11.9%) samples contained human Plasmodium parasites, and this is the first data report and further study is therefore needed.
Previous studies found Plasmodium relictum in avian species,19 P. cephalophi in ungulates,20 P. traguli in mousedeer,21 P. brucei in gray duiker,22,23 P. bubalis in water buffalo,24 and P. odocoilei in white-tailed deer.25,26 Other parasites found included P. caprae in goats (ruminant),27 P. bergei in Rodentia,28 and P. cynomolgi, P. inui, and P. fragile in primates.29 The five Plasmodium species that infect humans were originally parasites in primates.1,3,6–9 In this study, P. falciparum was found in buffalos, goats, dogs, and horses, while P. vivax was in buffalos, goats, and dogs. Initially, the presence of Plasmodium in these animals’ erythrocytes was not certain. However, the nested PCR showed the same results for all positive samples. The sequencing results of the positive bands in the nested PCR two analysis showed the bands were P. falciparum and P. vivax (Figure 3). This is the first investigation reporting human Plasmodium in domestic animals (ruminant, ungulate, and carnivore).
Plasmodium discovery among domestic animals in malaria-endemic areas raises the following questions. How do P. falciparum and P. vivax live in these animals? Are they intermediate hosts for this parasite? Did these Plasmodium species evolve to live in ruminants, ungulates, and carnivores? As a result of repeated exposure, have these animals become more permissive to Plasmodium, which generally lives in humans? Is this parasite pathogenic in animals? P. knowlesi is a commensal microbe in primates but pathogenic in humans1–3 and its migration from primates to humans is caused by forest loss or human invasion of primate habitat.11 There is a possibility that animal and human proximity aids easier cross parasite transfer between both groups by mosquitoes.
Despite high API in Fakfak and Gaura village, West Sumba, only the animals from West Sumba had human Plasmodium. This difference is possibly due to the distance between the residents’ houses and animal enclosures as the enclosures are located approximately 50–500 meters from the main houses in Fakfak. Meanwhile, in Gaura, residents live in stilt houses where the ground floor functions as an animal shelter, allowing microbial transfer between humans and animals by mosquitoes. In Fakfak, the sampling locations were not easily accessible, and the steep geographical conditions made it difficult to collect many samples compared to Gaura.
Although Plasmodium can be detected microscopically due to erythrocyte size, which is smaller in animals than humans, molecular methods become significant in detecting Plasmodium presence. The nested PCR was used to detect Plasmodium because its sensitivity was equally as high as Real-Time PCR and the cost was relatively lower.30,31 The microscopic method of double fluorescent dye utilization with Giemsa stain is recommended for further studies.32
In this study we found human Plasmodium in domestic animals. It is still not clear whether the animal had malaria, but this finding may be used as a reference for conducting malaria surveys in domestic animals in endemic areas. Human Plasmodium was only found in Gaura where the location of the animal enclosures is integrated with the residents’ houses. Local communities need to be educated about the possibility of malaria transmission due to the integration of animal enclosures and peoples’ homes. The discovery of human Plasmodium in domestic animals in this study may partly explain the persistence of the high prevalence of malaria in some endemic areas.
The author acknowledges the assistance of the West Sumba District and Fakfak Animal Husbandry Service, who helped carry out animal blood collection. We thank the people of Gaura village and Fakfak, who allowed us to take blood samples from their animals. We also thank Syahruni and Handayani Halik from Hasanuddin University Medical Research Center (HUM-RC) for their help with molecular work.
Figshare: Underlying data for ‘The discovery of human Plasmodium among domestic animals in West Sumba and Fakfak, Indonesia’, https://doi.org/10.6084/m9.figshare.14703012.v3.18
This project contains the following underlying data:
Figshare: ARRIVE checklist for ‘The discovery of human Plasmodium among domestic animals in West Sumba and Fakfak, Indonesia’, https://doi.org/10.6084/m9.figshare.14703012.v3.18
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Antimalarial drug and diagnostic resistance, molecular biology, molecular epidemiology, malaria elimination and control
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
No
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Malaria, genomics, genetics, immunology, epidemiology, invasion, immunology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
|
Version 4 (revision) 10 Dec 24 |
read | |||
|
Version 3 (revision) 09 Apr 24 |
read | read | ||
|
Version 2 (revision) 15 Feb 23 |
read | |||
|
Version 1 23 Jul 21 |
read | read | ||
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)