Keywords
LncRNAs, circadian rhythm, antisense, rhythmicity, feeding regimen
This article is included in the Cell & Molecular Biology gateway.
This article is included in the Genomics and Genetics gateway.
This article is included in the Circadian Clocks in Health and Disease collection.
LncRNAs, circadian rhythm, antisense, rhythmicity, feeding regimen
We revised our manuscript to clarify some points raised by the reviewers. We also added an extended data file that contains RNA expression levels of all the core clock genes (Table S2.xlsx, doi:10.6084/m9.figshare.21375783).
See the authors' detailed response to the review by Yao Cai
See the authors' detailed response to the review by Hikari Yoshitane
Long non-coding RNAs (lncRNAs) are a subgroup of RNAs longer than 200 nucleotides that do not produce proteins and play a variety of roles in a number of biological processes, including innate immune response,1 cell cycle control,2 cell differentiation,3,4 X-inactivation,5,6 and neuronal activity.7,8 Because lncRNAs do not produce proteins, it is important for lncRNAs to interact with other molecules, such as other nucleic acids and RNA-binding proteins, to exert their functions. For example, nuclear lncRNAs interact with DNA, chromatin, and proteins and modulate nuclear processes, like chromatin organization, RNA transcription, splicing, and lncRNA nuclear export and retention. In contrast, cytoplasmic lncRNAs interact with proteins, other RNAs, or different organelles to alter mRNA stability and localization, protein translation, post-translational modification, mitochondrial functions, or protein trafficking.9–15 In some cases, the act of transcription, rather than the lncRNA transcripts, is the functional entity to regulate target gene expression locally.16
Although significant progress has been made in understanding the functions of lncRNAs in recent years, their transcription regulatory mechanism has been poorly understood. Similar to mRNAs which are transcribed by RNA Polymerase II, a considerable number of lncRNAs are also transcribed by RNA Polymerase II, 5′ capped, 3′ polyadenylated, and multi-exonic.17 Interestingly, however, lncRNAs exhibit more cell type-, tissue-, developmental stage- or disease state-specific expression patterns compared to mRNAs.18–21 This has raised many interesting questions: What are the regulatory mechanisms of lncRNA transcription to achieve highly specific expression patterns? Is the transcription of lncRNAs also regulated by transcription factors which bind to its promoter/enhancer, similar to mRNAs? If so, why is lncRNA expression more specific? Is there a universal mechanism to regulate the transcription of all lncRNAs, or is the transcription of different lncRNAs regulated by different mechanisms? Does the expression of lncRNAs respond to external inputs similar to mRNAs?
We have recently shown that Per2AS, a lncRNA, plays an important role in regulating circadian rhythms,22 an internal timing mechanism to anticipate and respond to daily environmental rhythms driven by the rotation of the Earth. Interestingly, Per2AS is transcribed from the antisense strand of Period 2 (Per2), one of the core clock genes essential for generating circadian rhythmicity, and its expression is rhythmic and antiphasic to Per2 mRNA.23–26 Most strikingly, we further demonstrated that the transcription of Per2AS, rather than its transcripts, is important for regulating circadian rhythms.22 These data prompted us to interrogate how rhythmic Per2AS transcription is regulated.
In this study, we analyzed the expression patterns of Per2AS under conditions in which the circadian clock was perturbed either genetically or environmentally. We used publicly available circadian transcriptomic datasets from mouse liver, where Per2AS is abundantly and rhythmically expressed.23–26 Our first hypothesis is that the rhythmic Per2AS transcription is regulated by rhythmic antiphasic transcription of Per2 by means of transcriptional interference, in which the transcription process on one strand suppresses the transcription process of the other strand.27–29 Our alternative hypothesis is that rhythmic Per2AS expression is driven by its own promoter, similar to mRNAs.30–33 If the former is true, we anticipate that changes in Per2 and Per2AS expression would always be antiphasic. If the latter, then we anticipate that a change in Per2AS expression would be independent of that of Per2. We also take into account the possibility that the two hypotheses are not mutually exclusive. Results from this study contribute to our mechanistic understanding of how circadian rhythm is regulated by Per2AS and, more broadly, how the transcription of antisense lncRNAs is regulated.
All the fastq files were obtained from NCBI SRA (GSE135898, GSE135875, GSE107787, GSE102072, GSE143528,34–37 except for PRJDB7789, which was obtained from DDBJ DRA (PRJDB7789).38 Fastq reads were mapped to the Ensembl mouse genome release 38 (mm10) using STAR 2.7.7a39 with outFilterScoreMinOverLRead = 0.3 and outFilterMatchNMinOverLRead = 0.3 options. The ‘condenseGenes’ option was also used to select the most abundant isoform of each gene. The mapped reads were quantified by HOMER (v 4.11.1)40 and normalized by the transcripts per million (TPM) option. We used the -sspe option for paired-end reads and the -strand – or -strand + option to quantify mapped reads in a strand-specific manner. The option -bp10 was used for GSE102072 and PRJDB7789 to filter alignments with mapQ smaller than 10. Per2AS expression was calculated from the antisense strand of the Per2 genomic region as it is not annotated in Mus musculus GRCm38.95 GTF or NCBI RefSeq mm10 GTF files.
We used the two-way analysis of variance (ANOVA) in Microsoft Excel to test for differences in RNA levels between the experimental groups (i.e., genotype, diet), except for the dataset GSE102072 in which some samples had only one biological replica. The rhythmicity of each RNA expression was assessed by MetaCycle,41 which integrates three algorithms, ARSER, JTK CYCLE, and Lomb-Scargle, to determine the p-value, Benjamini-Hochberg q-value (BH.Q value), period, phase, baseline value, amplitude (AMP), and relative amplitude (rAMP). We defined the expression of an RNA as rhythmic when meta2d p<0.05.
To test whether any of the core clock genes have an effect on the expression of Per2AS and Per2, we analyzed the circadian transcriptomic datasets from mice livers lacking one or more “core” clock genes (Figure 1). Circadian rhythms in each cell are driven by a cell-autonomous molecular clock, composed of a group of core clock genes that form a network of transcriptional–translational feedback loops (TTFLs)42 and regulate daily rhythms in biochemistry, physiology, and behavior. In the first loop, transcription activators BMAL1 (gene name: Arntl) and CLOCK form a heterodimeric complex and bind to the E-box sequence in the promoter regions to regulate the rhythmic expression of their target genes, including Per1-3 and Cryptochrome (Cry)1-2. High levels of PER and CRY proteins form a protein complex in the cytoplasm, then translocate back to the nucleus to inhibit its own transcription by interacting with CLOCK/BMAL1. This cycle takes approximately 24 hours, and this is the molecular basis of generating circadian rhythms. The secondary feedback loop is comprised of the transcriptional repressors REV-ERBs (gene name: Nr1d) and the activator RORs that are regulated by BMAL1/CLOCK. REV-ERB and ROR proteins both recognize and bind to the RORE sequence in the promoter region and compete with each other to drive the rhythmicity of the target gene expression, including Bmal1. The last loop consists of transcription activators of proline and acidic amino acid-rich basic leucine zipper (PAR bZip) proteins: DBP, TEF, and HLF, and the repressor NFIL3, all of which target genes containing D-box element within their promoters, including Rev-erbs, Rors, and Pers.42
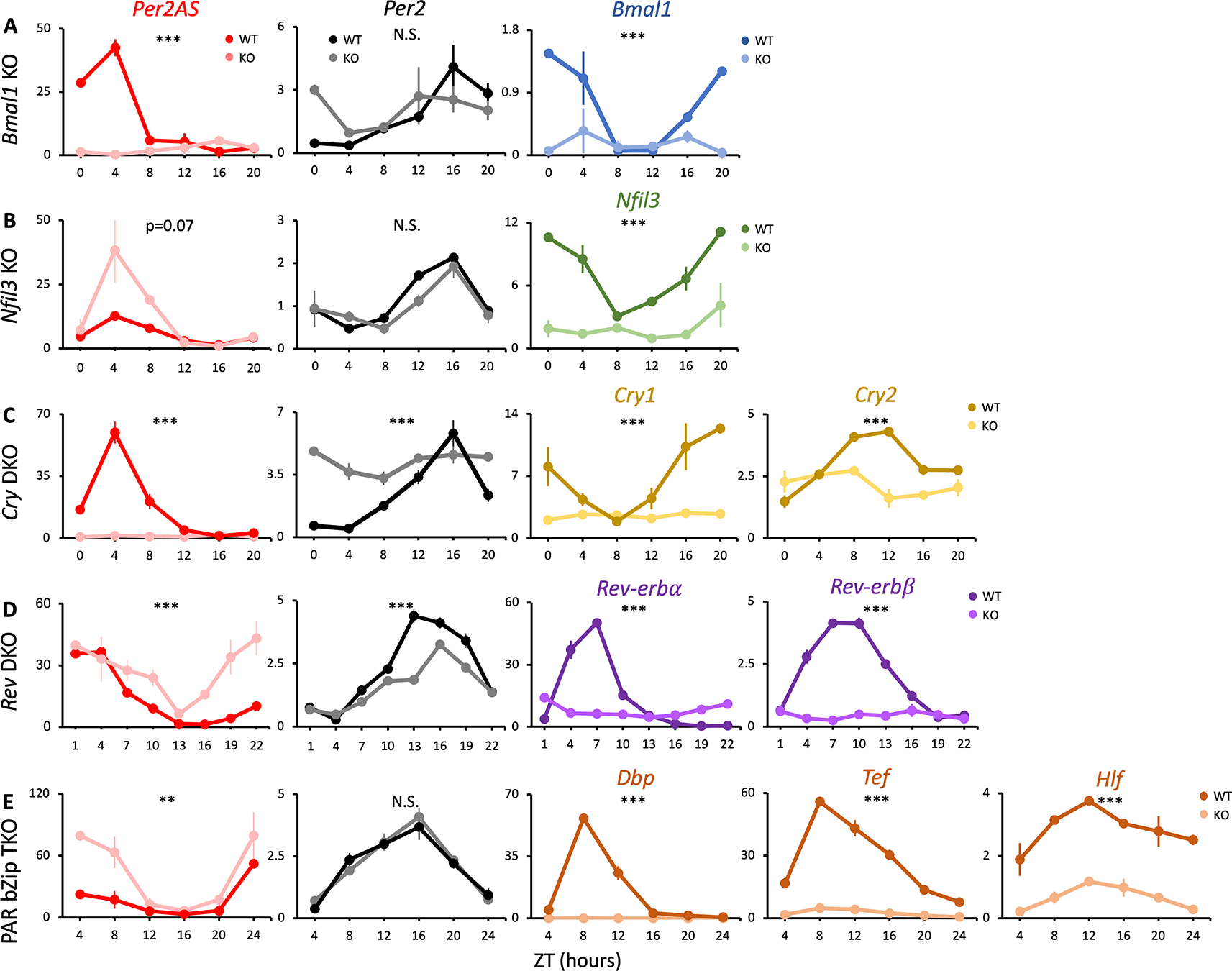
Mice were kept under an L:D=12:12 cycle and fed ad libitum. Liver RNAs were extracted every four hours in (A) Bmal1, (B) Nfil3, (C) Cry1/2 double, and (E) Dbp/Tef/Hlf triple knockout mice (n=2), or every three hours in (D) Nr1d1/2 double knockout mice (n=3). Y-axis represents strand-specific TPM. Points and error bars represent mean±SE. (**) p<0.01, (***), p<0.001, N.S. indicates no significant difference between genotypes (two-way ANOVA). Data derived from Weger et al., Proc Natl Acad Sci U S A, 202034; Yoshitane et al., Commun Biol., 201938; Guan et al., Science, 2020.37
Removal of Bmal1 from the TTFL results in arrhythmic locomotor activity and disrupts circadian expression of hepatic core clock genes.43,44 Similarly, removal of both Cry1 and 2 also results in the loss of the rhythm in locomotor activities and core clock gene expression.45,46 In both Bmal1 knock-out (KO) and Cry1/2 double knock-out (DKO) animals, our analysis demonstrated that the rhythmicity of Per2 was abolished but expression was maintained at intermediate levels, as was reported previously34,47,48 (Figure 1A, C, Table S185). Whereas the expression of Per2AS was markedly low and arrhythmic, indicating that Bmal1 and Cry1/2 promote the expression of Per2AS (Figure 1A, C, Table S185).
In contrast, the removal of Nfil3 or PAR bZip genes (Dbp/Tef/Hlf) did not change the expression pattern of Per2 (Figure 1B, E). This is in line with previous findings that the rhythmic expression of core clock genes was nearly identical between wild-type (WT) and Nfil3 or PAR bZip-deficient mice in liver.38,49 Interestingly, however, the amplitude of Per2AS increased in both Nfil3 KO and Dbp/Tef/Hlf triple knock-out (TKO) mice, indicating that the NFIL3 and PAR bZip proteins repress the expression of Per2AS without affecting Per2 (Figure 1B, E, Table S185).
Removal of both Nr1d1 and Nr1d2 in the SCN did not disrupt the circadian locomotor activity, but significantly shortened the free-running period.50 Also, Nr1d1/2 DKO mice showed interfered circadian expression of many hepatic core clock genes, including dampening the rhythm of Per2.51 Consistent with this, the amplitude of Per2 expression was decreased in Nr1d1/2 DKO mice while the relative amplitude of Per2AS expression was increased (Figure 1D, Table S185). These data suggest that Nr1d1/2 have an opposing effect on Per2 and Per2AS, activating Per2 while repressing Per2AS. We confirmed the genotypes of each dataset by checking the mRNA levels of the knock-out genes, all of which were significantly decreased (Figures 1, 3, 4). We also provided the mRNA levels of all the other core clock genes (Table S2).86
We next tested the effect of environmental perturbation on the expression patterns of Per2AS and Per2. In mammals, light is the most potent environmental cue that entrains the circadian clock through the suprachiasmatic nucleus (SCN) of the hypothalamus. However, food intake also serves as a strong ‘Zeitgeber’ (time giver) to entrain the circadian clock of peripheral organs in an SCN-independent manner.52–55 Previous studies have demonstrated that fasting directly affects a large number of physiological parameters, such as body temperature,56,57 body weight,58,59 hormone levels,60,61 and hepatic glucose levels,62,63 in addition to the expression patterns of the core clock genes in mouse liver.
In particular, the expression levels of BMAL1-target genes, including Per2, are lower in the liver of fasting mice.35,64–68 In line with this, the amplitude of Per2 expression was considerably lower in the fasted mice (Figure 2, Table S185) whose liver samples were collected after 24 hours of fasting for each time point, compared to the mice fed under the ad libitum (ad lib) condition. In contrast, the expression patterns of Per2AS show little or no difference between the ad lib fed and 24-hr fasting conditions (Figure 2, Table S185). These data indicate that the 24-hr fasting alters the expression pattern of Per2, but not that of Per2AS.
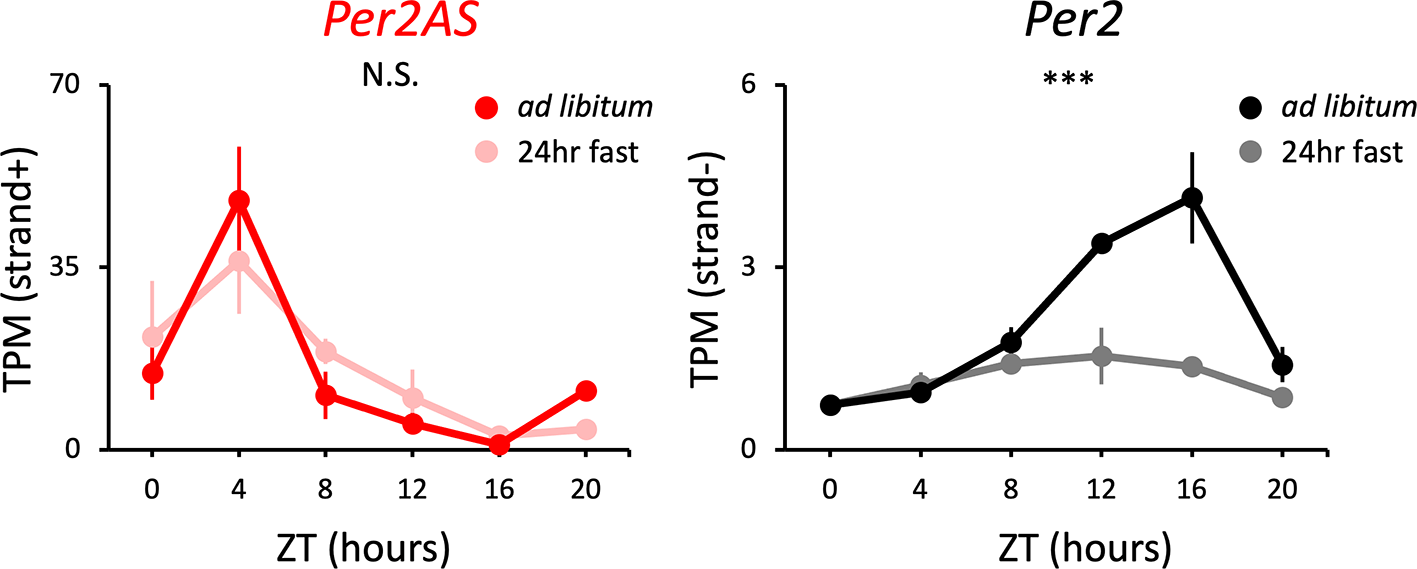
Mice were kept under an L:D=12:12 cycle and either fed ad libitum or food was removed 24 hours prior to tissue sampling for the fasting group of mice. Liver RNAs were extracted every four hours. Points and error bars represent mean±SE (n=3). (***) p<0.001, N.S. indicates no significant difference between feeding conditions (two-way ANOVA). Data derived from Kinouchi et al., Cell Rep., 2018.35
Time-restricted feeding (TRF) is a form of intermittent fasting in which food consumption is restricted to a certain time window of the day. The TRF regimen can protect mice from excessive body weight gain and liver damage depending on the timing of the food availability, and also improve metabolic and physiological rhythms when food access is restricted to the active phase.69–72 Here, we also looked at the differences in Per2AS and Per2 expression patterns when food access was restricted to nighttime (i.e., the active phase of mice) to understand whether core clock genes still have the same effect on the expression of Per2AS and Per2. Similar to what was observed with the ad lib feeding condition (Figure 1A, C), the expression of Per2AS was very low and arrhythmic in Bmal1 KO and Cry1/2 DKO animals experiencing TRF compared to WT (Figure 3, Table S185). The expression of Per2 was also arrhythmic but still maintained intermediate levels both in Bmal1 KO and Cry1/2 DKO animals even under TRF (Figure 3, Table S185). Compared to ad lib feeding, TRF did not affect the expression patterns of Per2AS and Per2, showing that the core clock gene KO is the primary factor to alter the expressions of Per2AS and Per2. Additionally, these findings further support that BMAL1 and CRY are crucial transcription factors to promote Per2AS expression, regardless of the feeding patterns.
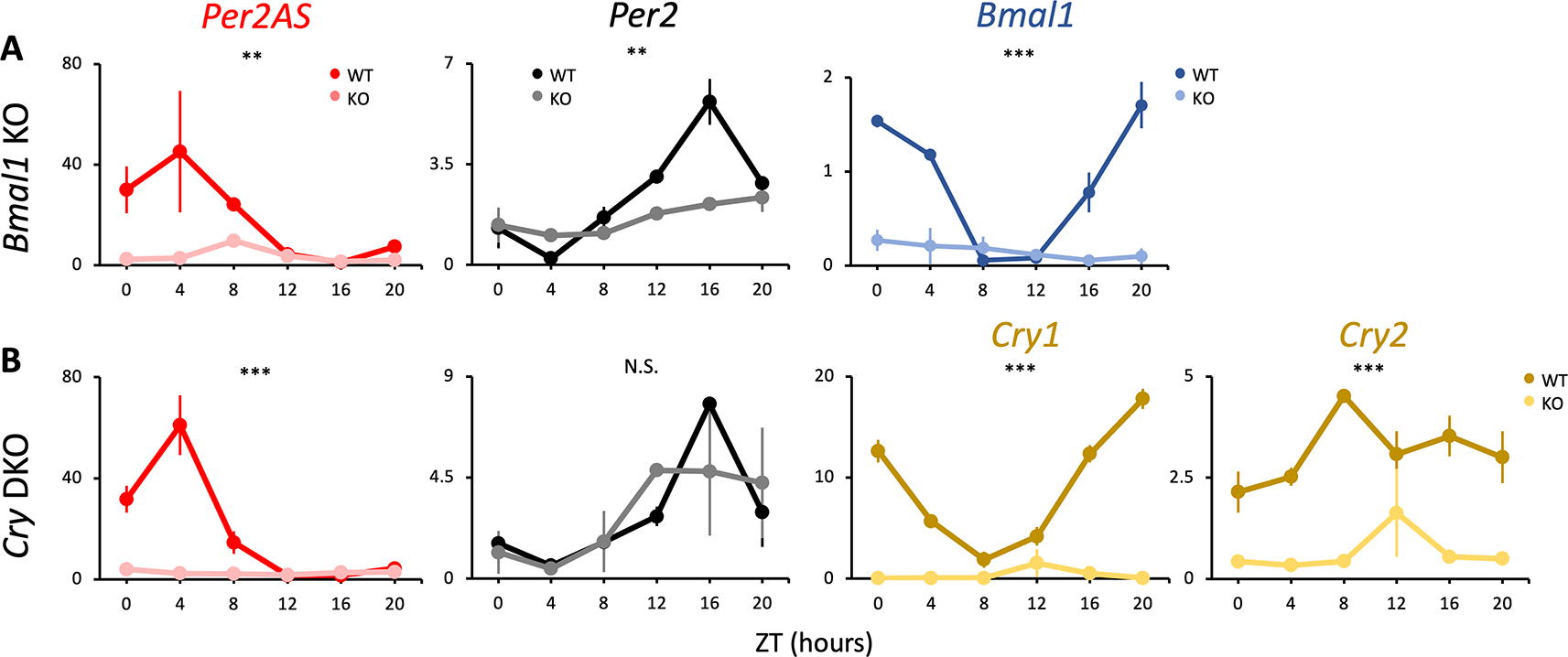
Mice were kept under an L:D=12:12 cycle and access to food were restricted between ZT12 to ZT24. Liver RNAs were extracted every four hours in (A) Bmal1 and (B) Cry1/2 double knockout mice. Y-axis represents strand-specific TPM. Points and error bars represent mean±SE (n=2). (**) p<0.01, (***) p<0.001, N.S. indicates no significant difference between genotypes (two-way ANOVA). Data derived from Weger et al., Proc Natl Acad Sci U S A, 2020.34
In addition to the timing of diet, the composition of the diet, such as high-fat or ketogenic, also affects circadian rhythms and alters core clock gene expression.69,73–76 To understand whether different diet compositions affect the expression of Per2AS and Per2 differently, we also examined the expression levels of Per2AS and Per2 when mice were fed with a 60% high-fat diet (HFD) either ad lib or TRF during the active phase (ZT13-22 or 23).36
The expression of Per2 was arrhythmic in WT fed with HFD under the ad lib condition (Figure 4A, Table S185), as was reported previously.74 However, it was rhythmic under the TRF condition (Figure 4D, Table S185), supporting the idea that TRF restores peripheral oscillations of core clock gene expressions.77,78 Similar to the results observed with a regular chow diet under the ad lib condition (Figure 1), Per2 expression was arrhythmic but maintained intermediate expression in both ad lib and TRF conditions in Bmal1 KO and Cry1/2 DKO (Figure 4, Table S185), but not in Nr1d1/2 DKO mice with HFD (Figure 4C, F, Table S185). In contrast, Per2AS levels were low and arrhythmic in both ad lib and TRF conditions in Bmal1 KO and Cry1/2 DKO mice compared to WT mice with HFD (Figure 4A-B, D-E), similar to what was observed in regular chow mice under both ad lib and TRF conditions (Figure 1A, C; Figure 3). This further supports the idea that Per2AS expression is regulated by Bmal1 and Cry1/2, regardless of the composition of the diet. On the other hand, Per2AS expression was arrhythmic in Nr1d1/2 DKO mice with HFD in contrast to the regular chow diet under ad lib condition (Figure 1D), suggesting that the effect of Nr1d1/2 on Per2AS is different between regular chow and HFD (Figure 4C, F; Table S185).
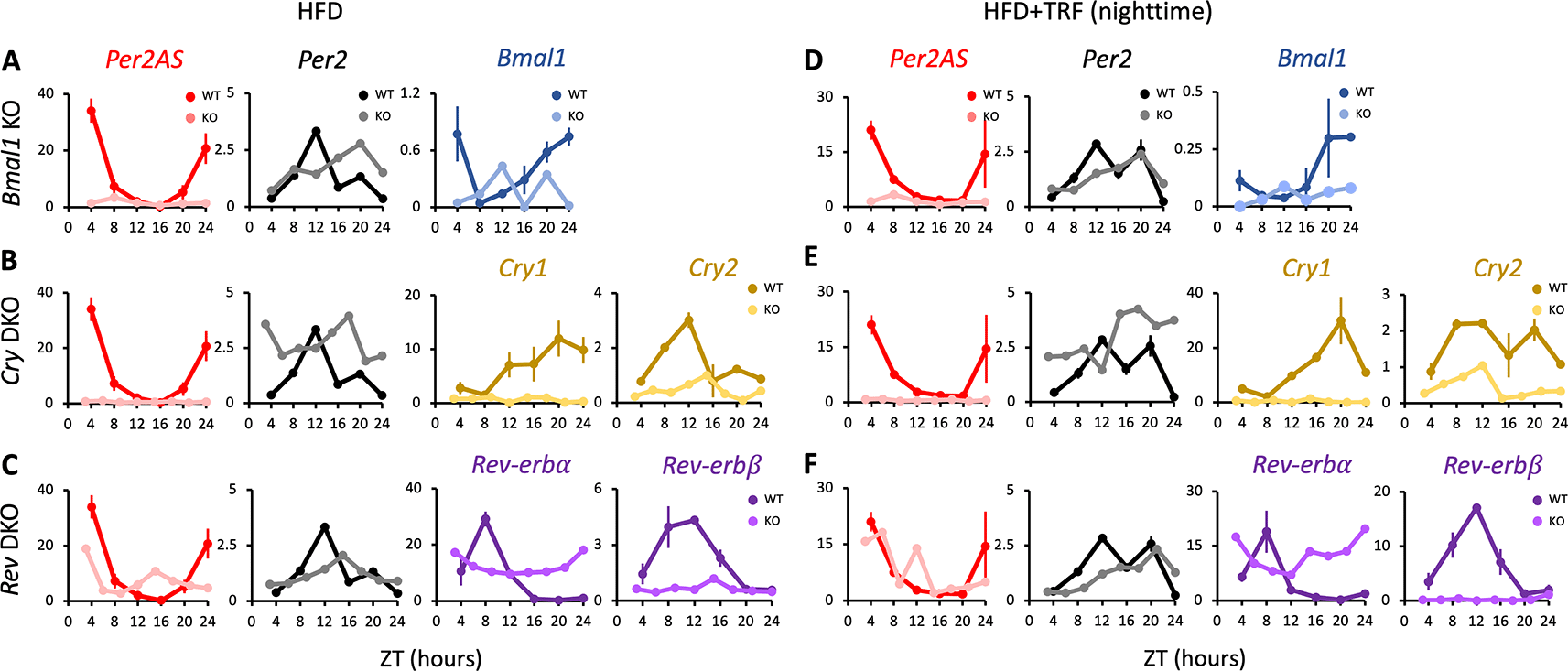
Mice were kept under an L:D=12:12 cycle and fed with a 60% high-fat diet either ad lib (A-C) or with TRF (D-F) condition. Liver RNAs were extracted every four hours in wild type and (A) Bmal1 knockout mice, or every three hours in (B) Cry1/2 double and (C) Nr1d1/2 double knockout mice. Y-axis represents strand-specific TPM. The same WT mice were used in each group. Points and error bars represent mean±SE (n=2). Two-way ANOVA analysis was not performed for this dataset, because the KO samples consisted of a single biological replicate. Data derived from Chaix et al., Cell Metab., 2019.36
In this study, we focused on the circadian antisense lncRNA, Per2AS, and asked whether the rhythmic expression of Per2AS is regulated independently by its own promoter like many other circadian mRNAs, or transcriptional interference driven by the antiphasic expression of its sense-strand gene Per2. By using circadian transcriptomic datasets from mouse liver, in which the molecular clock machinery was genetically and environmentally disrupted, we examined how these perturbations affect the expression patterns of Per2AS and Per2. Our data demonstrated that the expression of Per2AS can be altered by both genetic and environmental perturbations of the circadian clock.
We were able to test the effect of Bmal1, Nfil3, Cry1/2, Nr1d1/2, and Dbp/Tef/Hlf, but not that of Rora/b/c as the circadian transcriptome dataset for Rora/c KO mouse liver is only available with a microarray platform and this does not allow us to quantify Per2AS levels.79 We also tested the effect of fasting, TRF, and an HFD on the expression patterns of Per2AS and Per2. Even though the expression of many core clock genes, including Per2, is affected by fasting (Figure 2) or HFD (Figure 4),35,36,64–68 Per2AS appears to be less sensitive to these changes and its expression remained rhythmic under fasting, TRF, or HFD conditions (Figures 2-4). These data suggest that the timing of food intake, the composition of the diet, and fasting are not the primary factor that regulates the rhythmic expression of Per2AS. By contrast, the effect of genetic perturbation within the circadian rhythm system is stronger than environmental perturbation on the expression Per2AS.
We tested our hypotheses for Per2AS transcription regulation by comparing the changes in the expression pattern between Per2AS and Per2. We found that, in some cases, the Per2AS expression pattern was altered even when that of Per2 was unaltered (Figure 1B, E). We also found, in other cases, that the Per2AS expression pattern was unaltered even when that of Per2 was altered (Figure 2). These data strongly support the independent hypothesis that the expression of Per2AS is regulated by its own promoter, and its transcription is independent from that of Per2. Indeed, the expression of many lncRNAs is controlled by their promoter or other DNA elements, such as enhancers.30,80 The majority of lncRNAs also contain highly conserved core promoter sequences and can be regulated by different transcription factors.81,82 In addition, the promoters of lncRNAs are evolutionarily conserved as much as that of mRNAs at least between humans and mice, even though their nucleotide sequences are less conserved than mRNAs. These data support the significance of promoter sequences in regulating lncRNA expression patterns.83 Our data also indicate that BMAL1 and CRY1/2 are the activators, and NFIL3 and PAR bZip proteins are the repressors of Per2AS (Figure 1, 3). However, the removal of one particular core clock gene may alter the expression of other core clock genes or their downstream genes.34,84 Thus, we cannot eliminate the possibility that the Per2AS expression is indirectly impacted by the change of the core clock gene circuit.
Although these data strongly support the independent hypothesis, we cannot completely reject the alternative hypothesis that Per2AS expression is regulated by transcriptional interference from Per2, since there are some instances where the changes in the expression pattern of Per2AS and Per2 can still be explained by the transcriptional interference hypothesis. For example, in Bmal1 KO and Cry1/2 DKO animals under any dietary conditions (i.e., ad lib, TRF, and HFD feeding), the Per2AS expression was completely abolished, and the Per2 expression pattern was also arrhythmic but stayed at the intermediate levels (Figures 1A, C, 3, 4D, E, Table S185). It is possible that the constant Per2 expression prevents the transcription of Per2AS on the other strand, leading to the decreased and arhythmic Per2AS expression. Additionally, in Nr1d1/2 DKO mice, Per2AS expression increased and its rhythmicity became more robust, while Per2 expression decreased and its rhythmicity was dampened (Figure 1D). This could be due to decreased Per2 transcription leading to the increased Per2AS transcription on the other strand. Therefore, the two alternative hypotheses can both be viable and coordinated together to regulate the rhythmic transcription of Per2AS. At the same time, these data can also be explained solely by the independent hypothesis, and additional experimental evidence will be required to distinguish these possibilities. For example, it would be helpful to understand whether these core clock proteins are indeed recruited to the Per2AS promoter or enhancer sequences. We can also modify the transcription of Per2 directly and test whether this would lead to changes in Per2AS expression patterns.
Regardless, these results help us better understand not only how the transcription of Per2AS is regulated, but also how Per2AS is wired with other core clock proteins in transcriptional-translational feedback loops to regulate circadian rhythms because the rhythmic transcription of Per2AS is important for its functions in regulating circadian rhythms. More broadly, our results also help us understand the transcription regulation mechanism of antisense lncRNAs.
NCBI GEO: Temporal profiles of gene expression in Cry1/2 KO, Bmal1 KO under night restricted feeding and ad libitum feeding regimen. Accession number: GSE135898, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135898.
NCBI GEO: Temporal profiles of hepatic gene expression in PAR bZip triple knockout mice. Accession number: GSE135875, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE135875.
NCBI GEO: Fasting Imparts a Switch to Alternative Circadian Transcriptional Pathways in Liver and Muscle. Accession number: GSE107787, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE107787.
NCBI GEO: Hepatic transcriptome by Next Generation Sequencing of WT and clock mutant mice fed a HFD ad libitum or time-restricted feeding. Accession number: GSE102072, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE102072.
NCBI GEO: The Hepatocyte Clock and Feeding Interdependently Control Chrono-Homeostasis of Multiple Liver Cell Types (RNA-seq). Accession number: GSE143524, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE143524.
NCBI BioProject: Transcriptome of mouse liver in Per2::Luc KI and E4bp4 KO/ Per2::Luc KI mice. Accession number: PRJDB7789, https://www.ncbi.nlm.nih.gov/bioproject?term=PRJDB7789&cmd=DetailsSearch.
figshare: Table_S1.xlsx. https://doi.org/10.6084/m9.figshare.21067537.v1. 85
figshare: Table_S2.xlsx. https://doi.org/10.6084/m9.figshare.21375783.v1. 86
This project contains the following underlying data:
‐ Table S1.xlsx (Metacycle analysis to determine whether the gene expression is rhythmic)
‐ Table S2.xlsx (RNA expression levels (strand-specific TPM) of the core clock genes)
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
The authors thank Dr. Nobuya Koike from the Kyoto Prefectural University of Medicine for technical assistance and the members of the Kojima laboratory for critical proofreading of the manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Circadian clock, phosphorylation, Drosophila genetics
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
References
1. Adlanmerini M, Krusen BM, Nguyen HCB, Teng CW, et al.: REV-ERB nuclear receptors in the suprachiasmatic nucleus control circadian period and restrict diet-induced obesity.Sci Adv. 2021; 7 (44): eabh2007 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Circadian clock, post-translational modifications, phosphorylation, LC-MS/MS
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 31 Oct 22 |
||
|
Version 1 20 Sep 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)