Keywords
Virulence factors, Uropathogenic Escherichia coli, genes, antibiotic resistance, multiplex PCR
This article is included in the Manipal Academy of Higher Education gateway.
This article is included in the Pathogens gateway.
Virulence factors, Uropathogenic Escherichia coli, genes, antibiotic resistance, multiplex PCR
Changes have been done in the first line of the abstract and introduction part as per reviewer suggestions. The references 1 and 2 have been updated. The journal's name have been added in references wherever found missing and updated the citation. In the methodology section, reference number 8 has been cited. References have been updated in serial order thereon. The second line of the results section has been modified. Included limitations of the study in the last paragraph of discussion•
The authors have updated the discussion and conclusion in the paper to explain why nitrofurantoin would be suggested for therapy. Mention of the statistical method used for the association of the virulence factors detected and the resistance pattern of antibiotics has been made in discussion. The authors have updated the background of the study based on previously published literature, to look into the common genes papC and iutA and other genes hly A and cnf1, for epidemiological and virulence characterisation. The rate of catheter associated UTI has been added in the results section.
See the authors' detailed response to the review by Vignesh Ramachandran
See the authors' detailed response to the review by Anusha Rohit
Background: Urinary tract infection (UTI) is one of the most prevalent bacterial infection in humans. Every year, globally about 150 million people are diagnosed with UTI.1 Escherichia coli, Klebsiella spp., Pseudomonas aeruginosa, Proteus mirabilis, Citrobacter spp., Staphylococcus spp., Enterococcus spp., are the most common species causing UTI. Escherichia coli is the most frequent pathogen in the human urinary tract and accounts for 75% of the UTI. 85% of the community acquired UTIs and 40% of the hospital acquired UTI are attributed to Escherichia coli.2
The term uropathogenic E. coli (UPEC) refers to the extraintestinal strains of E. coli that express a variety of virulence factors, contributing to their pathogenicity in comparison to commensal E. coli.3,4 The specific virulence genes present in strains of UPEC isolates are the genes that encode adhesins (e.g. Type I fimbriae and P fimbriae), mechanisms for acquisition of nutrients (e.g. siderophores), factors that help the UPEC to escape from host defence systems (e.g. lipopolysaccharide, capsule) and toxins (e.g., cytotoxic necrotising factor 1, hemolysin).3 The factors mentioned above equip the bacteria with the ability to colonize the periurethral region, ascend the urinary tract to reach the urinary bladder resulting in infections like cystitis, urethritis, pyelonephritis, and urosepsis.5 UPEC strains have acquired the virulence genes (chromosomal or plasmid mediated by horizontal transfer of DNA). The molecular based platforms aid in the detection and characterization of UPEC strains.4 The characterization of these virulence genes in UPEC strains will help in better understanding of pathogenesis and course of UTI. Based on previously published literature,3,4 we aimed to look into the common genes papC and iutA and apart from them, other genes hly A and cnf1 were screened for epidemiological and virulence characterisation.
Recently there is an upsurge in UPEC strains that are multidrug-resistant (MDR), i.e., resistant to at least three or more classes of antibiotic agents.6 The emergence of MDR-associated UTI is increasing off late. Extended Spectrum β-Lactamase (ESBL) production among UPEC strains pose a therapeutic challenge. As a result, the therapeutic options available for the treatment of UTI are cut down which in turn is linked to treatment failure and increase in the economic burden of the community.7
Objectives: The study was undertaken to monitor the distribution of virulence factors among UPEC strains, to note the antibiogram, outcome, type of UTI and to look for the association of genetic virulence traits with antibiotic resistance.
Study design: Prospective cross sectional time bound study.
Study setting: The study was conducted in the Department of Microbiology, in a tertiary care center at Mangalore, India, for a duration of six months (Study period: December 2020 to May 2021).
Participants: The inclusion and exclusion criteria are as mentioned below:
Inclusion criteria: All clinically significant isolates of E.coli from urine. An isolate was considered significant if urine cultures had colony count ≥105 CFU/ml or ≥103 CFU/ml in symptomatic patients.1
Exclusion criteria: Urine samples with no growth, less significant counts of E.coli, growth of bacteria other than E.coli, isolates of E.coli from clinical samples other than urine.
The study was conducted after approval from the Institutional Ethics Committee, Kasturba Medical College, Mangaluru (Reg No. ECR/541/Inst/KA/2014/RR-17) Reference number IECKMCMLR-12/2020/408). The study was performed on isolates retrieved in the laboratory obviating the need for informed consent from the patients.
Specimen processing: Clean catch midstream urine samples received from suspected cases of UTI were processed within one hour of collection. The samples were inoculated onto MacConkey’s agar by semi quantitative method, Cystine-Lactose-electrolyte-deficient agar, UTI chrome agar. The culture plates were incubated for 37°C for 24 hrs. Urine samples with pure growth of E.coli, with a colony count of ≥105 CFU/ml or ≥103 CFU/ml in symptomatic patients, were considered significant. The study included 75 urinary isolates of E. coli. The isolates were identified based on colony morphology and standard biochemical tests. Antibiotic susceptibility testing was performed by the modified Kirby–Bauer disk diffusion/Vitek2 Compact (Biomerieux, France) system and interpretation was done as per the Clinical and Laboratory Standards Institute guidelines.8
Variables and data source: The phenotypic methods for haemolysin production, serum resistance and genotypic characterization of virulence genes in UPEC are explained below.
Phenotypic methods for detection of haemolysin production and serum resistance:
The detection of α-haemolysin produced by E. coli was performed by plate haemolysis test. The presence of a zone of complete lysis of erythrocytes around the colony and clearing of the medium on 5% sheep blood agar, is suggestive of α-haemolysin production.9,10
Serum resistance was studied by using fresh overnight culture of isolates as per the method described by Sharma et al.10 The UPEC strains were considered serum sensitive if viable count dropped to 1% of the initial value and serum resistant if ≥90% of organisms survived after 180 minutes.
Genotypic characterization of virulence genes of UPEC:3,11
DNA extraction was performed by boiling method. The spectrum of virulence genes in UPEC strains was detected using two sets of multiplex PCR as shown below. Table 1 shows the PCR mastermix preparation used. The primer sequence of the mentioned genes is shown in Table 2. PCR was performed in a final reaction volume of 50 μl. The program for amplification included a step of initial denaturation at 95°C for 3 min, followed by 25 cycles of 94°C for 30 s, 61°C for 30 s and 68°C for 3 min and a final extension step at 72°C for 3 min. The amplicons are visualized using the gel documentation system.
The required data was retrieved from the clinical case records and the cases of complicated and uncomplicated UTI were identified. Uncomplicated UTIs are those that occur in healthy individuals without any of the predisposing factors for UTI. Complicated UTIs occur in individuals with underlying functional or structural abnormalities of the genitourinary tract.12
Sample size: The sample size was calculated taking into account the data of the previously published article (2). Using the formula , a sample size of total 75 is calculated. Where p=prevalence=75%, q=1-p, d=Effect size=10%, Z=1.96 at 80-95% confidence interval.
The sample size for the study was 75.
All the data was entered into an excel sheet and analyzed using IBM SPSS version 25. The continuous and categorical variables have been represented as mean ± standard deviation and frequency percentages respectively. The association between the variables were analyzed using the chi-square test.
A total of 75 urinary isolates of E.coli from patients with suspected urinary tract infections were included. Females had a higher preponderance of UTI (66.7%) than males (33.3%). Only 5 (7%) of the 75 patients were paediatric patients, remaining 70 (93%) were adults. The majority of the female patients were in the age group 20-39 years.
Phenotypic detection of serum resistance and hemolysin production exhibited by E.coli was 100% and 32% respectively. Multiplex PCR was performed for the detection of virulence genes papC, Cnf1, hlyA & iutA as shown in Figures 1 and 2. The distribution of virulence genes among the 75 UPEC isolates is as shown in Figure 3. Out of 75 isolates, 65 were positive for at least one of the four targeted genes as shown in Table 3, while the remaining ten isolates were negative for all 4 genes.
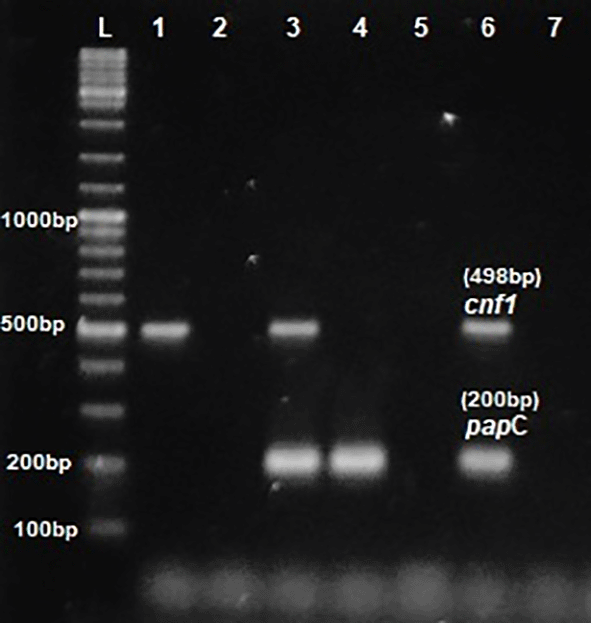
L- ladder 1000+ bp, lane 1-6: test isolates, lane 7: Negative control.
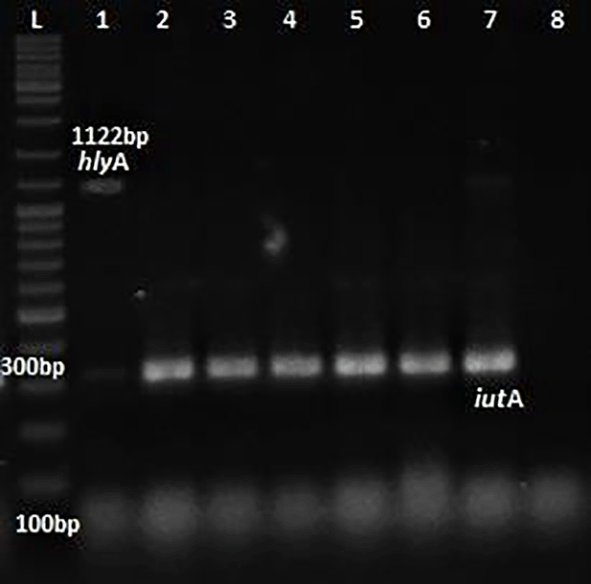
L: ladder 1000+ bp, lane 1-7: test isolates, lane 8: Negative control.
It was found that hlyA gene was found in a higher percentage in haemolytic isolates (41.7%) than non- haemolytic isolates (19.6%). p-value was statistically significant (p-value=0.044). Also, 50% of the cnf1 positive isolates harboured the hlyA (haemolysin) gene (p=0.003; statistically significant).
The antibiotic resistance pattern of the UPEC isolates are as shown in Figure 4. Out of 75 isolates, 38 (50.6%) isolates were ESBL producers. Multidrug resistance (resistance to three or more antibiotic classes) was found in 40 (53.3%) isolates. Our study revealed that 45 out of the 75 urinary isolates, possessed more than one virulence factor as shown in Table 5.
The distribution of virulence factors in antibiotic-resistant isolates were studied as shown in Table 4. Statistical analysis (Chi square test ) for the association of the virulence factors detected and the resistance patterns of the isolates was done. Hemolysin production and resistance to imipenem, Norfloxacin was found to be significant (p≤0.05). The presence of hlyA gene and resistance to ceftazidime was found to be significant (p≤0.05). There was no statistically significant difference between the presence and absence of the other virulence genes with specific antibiotic resistance.
| Virulence factors and antibiotic resistance | A | PiT | Ctr | Caz | Cfs | I | Etp | Mer | G | Ak | Nx | Cip | Fos | Nit | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haemolysin | +ve (24) | 61.9% (21) | 22.7% (5) | 56.5% (13) | 47.8% (11) | 4.3% (1) | 12.5%* (2) | 4.8% (1) | - | 21.7% (5) | 4.2% (1) | 19% (4)* | 39.1% (9)* | 4.2% (1) | 4.3% (1) |
| -ve (51) | 80.9% (38) | 10% (5) | 66.7% (32) | 34% (17) | 2.1% (1) | - | - | 2.1% (1) | 18% (9) | - | 61.7% (29)* | 74% (37)* | - | 2% (1) | |
| SR | +ve (75) | 75% (51) | 13.9% (10) | 63.4% (45) | 38.4% (28) | 2.9% (2) | 4.3% (2) | 1.4% (1) | 2.1% (1) | 19.2% (14) | 1.4% (1) | 48.5% (33) | 63% (46) | 1.4% (1) | 2.7% (2) |
| papC | +ve | 70.6% (24) | 10.8% (4) | 64.9% (24) | 45.9% (17) | 2.7% (1) | - | - | 3.8% (1) | 13.2% (5) | - | 40% (14) | 51.4% (19) | - | 2.6% (1) |
| -ve | 79.4% (27) | 19.1% (6) | 61.8% (21) | 30.6% (11) | 3% (1) | 8.7% (2) | 3% (1) | - | 25.7% (9) | 2.9% (1) | 57.6% (19) | 75% (27) | 2.9% (1) | 2.9% (1) | |
| Cnf1 | +ve | 76.2% (16) | 19% (4) | 68.2% (15) | 47.6% (10) | - | - | - | - | 27.3% (6) | 33.3% (7) | 47.6% (10) | - | - | |
| -ve | 74.5% (35) | 11.8% (6) | 61.2% (30) | 34.6% (18) | 4% (2) | 6.1% (2) | 2% (1) | 3% (1) | 15.7% (8) | 2% (1) | 55.3% (26) | 69.2% (56) | 2% (1) | 3.9% (2) | |
| hlyA | +ve | 73.3% (11) | 73.7% (14) | 66.7% (12) | 73.7% (14)* | 5.3% (1) | - | - | - | 26.3% (5) | - | 33.3% (5) | 63.2% (12) | 5% (1) | - |
| -ve | 75.5% (40) | 84.9% (45) | 62.3% (33) | 25.9% (14)* | 2% (1) | 6% (2) | 2% (1) | 2.8% (1) | 16.7% (9) | 1.9% (1) | 52.8% (28) | 63% (34) | - | 3.8% (2) | |
| iutA | +ve | 81.4% (35) | 14.9% (7) | 66.7% (30) | 38.3% (18) | 2.1% (1) | - | - | 3.2% (1) | 21.3% (10) | - | 57.1% (24) | 63.8% (30) | 2.1% (1) | - |
| -ve | 64% (16) | 12% (3) | 57.7% (15) | 38.5% (10) | 4.3% (1) | 15.4% (2) | 4.2% (1) | - | 15.4% (4) | 3.8% (1) | 34.6% (9) | 61.5% (16) | - | 7.7% (2) | |
Among the 40 MDR E. coli isolates, 11 (27.5%) isolates produced haemolysis and all were serum resistant. iutA, papC, cnf1, hlyA genes were detected in 72.5% (n=29), 47.5% (n=19), 35% (n=14) and 27.5% (n=11) of the MDR isolates respectively.
Out of the total 75 cases of UPEC studied, 60% (n=45) were complicated UTI and 40% (n=30) were uncomplicated UTI. Out of the complicated UTI cases, the common genes detected were iutA (83%) and papC (50%) followed by cnf1 (33%) and hlyA (10%) genes. Among the uncomplicated UTI isolates, iutA (77%), papC (55%) and cnf1 (23%) were the common genes detected. None of the isolates from uncomplicated UTI had hlyA. 77% of the complicated UTI were MDR and 33% of the uncomplicated UTI cases were MDR.
Out of the total 75 cases of UPEC studied, majority (97.4%) had a favourable clinical outcome at discharge. Catheter associated UTI was seen in 33.3% (n=25) of the patients. Mortality due to urosepsis was noticed in two cases (2.6%).
Urinary tract infection is one of the infectious diseases which is most prevalent amongst the people of all age groups from neonate to geriatric age group. Studies from India have reported varying prevalence rates of E.coli associated UTI- 50%-, 75%.13 The bacterial pathogen accounting for the majority of community and hospital-acquired urinary infections is the UPEC. Depending on the virulence, these infections might range from mild uncomplicated UTI to complicated UTI.2 Characterization of UPEC isolates with respect to their antibiotic resistance patterns and virulence factors, will aid in the effective management of UTI.
Our study revealed that 32% of the isolates produced haemolysin which is similar to the findings of Chhaya shah et al 34%,14 but lower compared to the findings of Shetty et al (60%).15 The exotoxin implicated in the enhanced virulence and lethality of clinical infections among UPEC strains is Alpha-hemolysin (Hly) production.16 UTIs associated with Alpha-hemolysin may be associated with extensive kidney inflammation and injury due to it’s cytotoxic nature. The majority of hlyA-positive strains were identified in patients with pyelonephritis (> 70%) compared to from patients with cystitis (31–48%), ascertaining the role of hlyA as an important virulence factor in the pathogenesis of pyelonephritis.17
Serum resistance is a property that provides a survival advantage to the bacteria. This makes UPEC resistant to killing by the alternative complement pathway in the normal human serum. Serum resistance in UPEC strains has been typically associated with pyelonephritis, cystitis and bacteraemia.7 In the current study, all 75 isolates showed resistance to serum bactericidal action. This is similar to the studies by Sharma et al and Shetty SK et al which reported 86.7% and 83% of serum resistance.10,18 However, study by Anuradha et al revealed 51% of serum resistance.7 In UPEC, capsule and the O antigen are polysaccharides that contribute to virulence. The extracellular polysaccharides are antiphagocytic and inhibit complement-mediated lysis.19 All the strains in this study on UPEC showed serum resistance indicating its significant association with UTI.
In the current study, the genes coding for adherence gene (papC), Cytotoxic necrotizing factor I (cnf1), α haemolysin (hlyA) and ferric aerobactin receptor (iutA) were detected by multiplex PCR.
The virulence gene iutA, is the gene for the siderophore ferric aerobactin receptor which helps the bacterial intake of iron and this enables the survival of bacteria in an atmosphere with limited concentrations of iron (urinary tract) This gene was the commonest gene detected in the UPEC isolates of our study(65%), thus proving its association with the virulence of UPEC. This finding supports the data published by Munkhdelger et al (62.2%),20 and Karam et al (66.4%).21
The rate of detection of adherence gene (papC) was 52%. This is in par with a study by Chakraborty A et al in 2017 in which nearly half of their isolates (49%) carried this gene.3 Firoozeh et al reported a lower rate of detection (34.6%) of papC gene in UPEC isolates.22
27% of our isolates were positive for hlyA gene which was similar to the study by Gohar et al (26%).23 A lower rate (13.6%) of hlyA gene were seen by Daniela A et al study.24
Toxigenic strains of E.coli like necrotoxigenic E.coli-1(NTEC-1) produce Cytotoxic necrotizing factor 1 (cnf1).25 Our study showed that 29%(n=33) were NTEC -1 strains harbouring the gene cnf1. The distribution of cnf1 in our study is similar to the studies by Chakraborty A et al (29.5%) and Gohar et al (30%).3,23 NTEC -1 strains are emerging pathogens in India.25 Our study revealed that 50% of the cnf1 positive isolates harboured hlyA (haemolysin) gene, which is statistically significant. The combined production of several powerful toxins (haemolysin, CNF) and co-expression of various virulence genes by NTEC strains makes them potentially aggressive pathogens.25 Hence identification of these strains at an early stage, would prevent the complications associated with NTEC -1 strains.
The virulence genes studied are involved in the pathogenesis of UTI. However the absence of genes in 10 isolates (13.3%) could possibly be due to mutation of the gene Thus, a negative PCR result doesnot rule out the absence of virulence genes.
Varying spectrum and rates of antibiotic resistance have been reported among the UPEC isolates and most studies have reported that amikacin, nitrofurantoin and imipenem are highly efficacious against such strains.21 In our study, a high percentage of resistance was noted to beta lactam group of antibiotics (ampicillin-75%, ceftriaxone-64.4%), fluoroquinolones (ciprofloxacin-63%, norfloxacin-50%), trimethoprim/sulfamethoxazole (49.3%). Lower rates of resistance was detected with respect to piperacillin/tazobactam(13.9%), carbapenems (ertapenem-1.4%, imipenem-6.4%), amikacin(2.7%) and nitrofurantoin (2.7%), Cefoperazone/sulbactam (2.9%) and fosfomycin (1.4%). Based on these findings, nitrofurantoin, piperacillin tazobactam and cefaperazone sulbactam maybe suitable candidates for empirical therapy of UTIs. However, the therapeutic utility of these drugs in complicated UTI in comparison with uncomplicated UTI was not studied. Drugs like aminoglycosides, carbapenems and fosfomycin maybe used as reserve drugs in the management of MDR-UTI due to E. coli.
Nitrofurantoin being an oral drug maybe a good drug of choice in such patients with UTI and outpatients. These results are in par with a study which revealed that nitrofurantoin and carbapenems are suitable as empirical antibiotics in the treatment of UTIs in outpatients and fosfomycin can be used against highly resistant UPEC isolates.26 However, their inappropriate usage may gradually give rise to their increase in antibiotic resistance. This accentuates the need for local hospital data compilation and antibiotic resistance analysis to devise a suitable antibiotic policy for the hospitals.
We found that 51% were ESBL producers which is similar to studies by Chhaya et al (46%) & Shoba et al (19%-59.6%).14,27 In our study, 45 isolates expressed multiple virulence factors (Table 5), among which a majority (57.7%) were non ESBL producers. These results support the fact that the expression of virulence genes maybe inversely related to the presence of antibiotic resistance and ESBL production.
| Multiple virulence factors (no. of isolates) | ESBL + | ESBL - |
|---|---|---|
| + (45) | 19 | 26 |
| _ (30) | 19 | 11 |
| Total 75 | 38 | 37 |
The distribution of virulence factors in antibiotic-resistant isolates showed that hemolysin production is significantly associated with resistance to imipenem, norfloxacin and ciprofloxacin (p≤0.05) at par with findings of Chhaya et al.14 The presence of hlyA gene is significantly associated with resistance to Ceftazidime (p≤0.05). The association of the presence of hlyA with resistance to third generation cephalosporins poses a therapeutic challenge.28 No other statistically significant difference was proved between the presence or absence of the other virulence factors, with any specific antibiotic resistance.
Multidrug resistance was observed in 40 isolates (53.3%). This finding is comparable to the study by Hasan et al29 in India, in which the prevalence of MDR E. coli was 52.9%. However, this finding is in contrast to study by Munkhdelger et al21 which revealed a higher rate of MDR in UPEC (93.9%).
In our study, 27.5% (n=11) MDR isolates produced haemolysis and all the 40 MDR isolates showed serum resistance. Co-existence of MDR with hemolysis or serum resistance may contribute to the increased pathogenicity and nonresponse to therapy in cases of UPEC associated UTI. In our study, iutA, papC, cnf1, hlyA genes were detected in 72.5% (n=29), 47.5% (n=19), 35% (n=14) and 27.5% (n=11) of the MDR isolates respectively. iutA was the commonest gene detected in MDR isolates. This finding supports the fact that certain virulence genes like iutA is positively associated with MDR.30
Inappropriate use of broad-spectrum antibiotics, prolonged hospitalisation, poor hygiene are few major factors that contribute to increasing MDR infections. In our research, the percentage of MDR in complicated UTI was 77%; which was higher compared to uncomplicated UTI (33%). Our study supports the findings of a study by Johnson J R et al, which revealed that antibiotic resistance was higher in people with complicated UTIs than with uncomplicated UTIs.31
In our study, third generation cephalosporins (n=15, 37.5%), meropenem (n=12, 30%), piperacillin tazobactam (n=8, 20%) and nitrofurantoin (n=5, 12.5%) were the antibiotics used in the treatment of these MDR cases.
Out of the total 75 cases of UPEC studied, majority i.e 60% (n=45) were cases of complicated UTI. Distribution of cnf1 gene was more in complicated UTIs compared to uncomplicated UTIs. hlyA gene detected in complicated UTIs was absent in isolates from uncomplicated UTIs. This difference in results, regarding the distribution of hlyA and cnf1 genes, maybe attributed to the fact hemolysin production and cnf1 are attributed to the dysfunction of local immune response and host tissue damage. Thus, probably there is increased expression of hlyA and cnf1 in complicated UTIs. There is no significant difference in the distribution of genes associated with adhesion (papC) and iron uptake (iutA). This finding is in contrast to the study by Takahashi et al, which revealed that the prevalence of papC was more in cases of uncomplicated UTI.32
The three isolates from patients with history of emphysematous pyelonephritis and urosepsis, were found to be positive for virulence genes papC (adherence) and cnf1 (cytotoxicity). cnf1 gene is associated with extensive tissue damage4 and this could have contributed to the emphysematous pyelonephritis in these three cases.
Mortality due urosepsis was noticed in two cases (2.6%). Isolates of both these cases had all the 4 genes, produced hemolysin, were serum resistant and MDR. This substantiates the contribution of hemolysin production, serum resistance and expression of the virulence genes in the increased pathogenicity of UPEC.
Limitations of the study were that the study could have been performed on a larger sample size for better characterization of UPEC. The other limitation would be that the authors could not compare the association of the comorbidities such as immunocompromised condition and exposure to frequent antibiotics to look for any differences in the antibiotic resistance patterns, since the virulence characteristics are different among those populations. One more limitation was that data on nosocomial/community acquired UTI was not studied.
The association of hemolysin production with resistance to imipenem and norfloxacin in UPEC strains was found to be significant and the presence of hlyA gene is positively associated with ceftazidime resistance, thus posing a therapeutic challenge. Nitrofurantoin, piperacillin tazobactam, cefoperazone/sulbactam, carbapenems, fosfomycin and amikacin were the antibiotics against UPEC which showed lower rates of resistance. Owing to the lower rate of resistance and ease of administration, nitrofurantoin maybe an effective oral drug in outpatients with UTI. While Piperacillin tazobactam and cefaperazone sulbactam maybe suitable for empirical therapy of UTIs in inpatients. Drugs like aminoglycosides, carbapenems and fosfomycin may be used as reserve drugs in the treatment of MDR-UTI. However, inappropriate usage can gradually increase antibiotic resistance. Hence, proper selection of antibiotics in hospitals taking into account the local antibiogram is needed to reduce the emergence of antibiotic resistance. The data on distribution of virulence factors and antibiotic resistance helps in planning the management strategies in UTI in our setup, thus improving patient care.
Dryad. Characterization of virulence factors and antibiotic resistance pattern of Uropathogenic Escherichia coli strains in a tertiary care center. The full reference for data repository provided by dryad for access is as follows: DOI: https://doi.org/10.5061/dryad.q83bk3jmd.33
1. Mr Naveen Kumar M: Conceptualization, Data Curation, Formal Analysis, Investigation, Methodology, SoftwareSupervision, Validation, Visualization, Writing & Editing
2. Dr Sevitha Bhat: Data Curation, Formal Analysis, Investigation, Methodology, Project Administration, Supervision, Validation, Visualization, Writing – Review & Editing
3. Dr Archana Bhat K - Conceptualization, Data Curation, Formal Analysis, Investigation, Methodology, ProjectAdministration, Resources, SoftwareSupervision, Validation, Visualization, Writing – Review & Editing
4. Dr Shalini Shenoy M: Conceptualization, Project Administration, Resources, Supervision, Writing – Review & Editing
5. Dr Vishwas Saralaya: Conceptualization, Project Administration, Resources, Supervision, Writing.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Medical Microbiology
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Antimicrobial resistance, infection control
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Medical Microbiology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 28 Nov 22 |
read | |
|
Version 1 11 Oct 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)