Keywords
Circadian clock, Neurospora cras, tRNA synthetases, translation control
This article is included in the Cell & Molecular Biology gateway.
This article is included in the Circadian Clocks in Health and Disease collection.
Circadian clock, Neurospora cras, tRNA synthetases, translation control
In the revised manuscript, ribo-seq heatmaps of all aaRS were added to Figure 1A to show both rhythmic and nonrhythmic ribosome occupancy. New control data showing that both Pasprs::luc and Pglnrs::luc are arrhythmic in clock mutant Δfrq cells were added to Figure 2 (D and E). The strains and additional data were generated and analyzed by Teresa M. Lamb and Madhusree Gangopadhyay, both of whom were added as authors. The data from Figure 4C examining GlnRS rhythms in Δadv-1 cells was reanalyzed to include only the time points from DD12 to DD96. These data fit to a sine wave with a period of 22.5 h, but with a low amplitude compared to WT. This information is now included in Table 2, and we clarify in the text that deletion of transcription factor ADV-1 decreased the amplitude of GlnRS rhythms. In the text we added background information on why particular transcription factors were chosen for this study, added an updated reference on what is known regarding the regulation of eukaryotic aaRS genes, and provided a brief description of published Ribo-seq datasets used in this study in the methods.
See the authors' detailed response to the review by Ruifeng Cao
See the authors' detailed response to the review by Luis F. Larrondo
Aminoacyl-tRNA synthetases (aaRSs) play a fundamental role in mRNA translation by catalyzing the attachment of specific amino acids onto their cognate tRNAs. For accuracy, aaRSs employ chemical selectivity and proofreading capabilities (Rubio Gomez and Ibba, 2020, Roux and Topisirovic, 2012). Mounting evidence supports that aaRSs have functions beyond their role in charging tRNAs, including roles in immune signaling, cell cycle, nutrient metabolism and growth, and thus are linked to various human diseases (Pang et al., 2014, Nie et al., 2019, Park et al., 2008). Aminoacylation mechanisms are conserved across all kingdoms of life. The regulation of aaRS expression in prokaryotes is well-described (Green et al., 2010, Pelchat and Lapointe, 1999), and while less is known about their regulation in eukaryotes, coordinate induction of several mammalian cytoplasmic aaRS genes by amino acid starvation has been observed (Shan et al., 2016). In addition, several aaRSs were reported to have daily rhythms in abundance at the mRNA and/or protein levels in the filamentous fungus Neurospora crassa (Sancar et al., 2015, Hurley et al., 2018, Castillo et al., 2022b), and mammalian cells (Pembroke et al., 2015, Barclay et al., 2012, Hughes et al., 2009, Miller et al., 2007, Vollmers et al., 2012, Eckel-Mahan and Sassone-Corsi, 2009, Geyfman et al., 2012, Yoshitane et al., 2014, Janich et al., 2015). These data suggested that the circadian clock imparts regulation on aars gene expression, which would impact rhythmic protein synthesis and clock-controlled cellular processes.
The circadian clock is an endogenous timekeeping mechanism that regulates diverse biological processes in many organisms, allowing them to anticipate and prepare for daily environmental cycles, and to organize cellular processes to the right time of day for improved fitness (Dunlap and Loros, 2017). Disruption of the circadian clock has profound effects on human physiology and behavior, and can lead to a wide range of diseases (Bass, 2017, Foster, 2020, Hernandez-Garcia et al., 2020). Depending on the organism and tissue type, the circadian clock regulates daily rhythms in mRNA and protein accumulation for up to 50% of the eukaryotic genome (Hurley et al., 2014, 2018, Zhang et al., 2014, Mauvoisin et al., 2014). Remarkably, most of the proteins that cycle in abundance under the control of the circadian clock are produced from mRNAs that are not clock-controlled (Reddy et al., 2006, Robles et al., 2014, Zhang et al., 2014, Hurley et al., 2018, Castillo et al., 2022b). These data suggested a prominent role for clock regulation of post-transcriptional processes, including rhythmic mRNA translation.
The N. crassa circadian clock is composed of negative elements FREQUENCY (FRQ), FRQ-INTERACTING RNA HELICASE (FRH), and CASEIN KINASE 1 (CK1), and positive elements WHITE COLLAR-1 (WC-1) and WHITE COLLAR-2 (WC-2) (Baker et al., 2012, Dunlap and Loros, 2017). WC-1 and WC-2 heterodimerize to form the White Collar Complex (WCC) which binds to the promoters of frq and downstream clock-controlled genes (ccgs), including 24 transcription factors, to drive their rhythmic transcription (Froehlich et al., 2003, Smith et al., 2010). In addition, the N. crassa clock generates rhythms in the activities of the conserved eukaryotic translation initiation factor 2 (eIF2) and eukaryotic translation elongation factor 2 (e-EF2) (Caster et al., 2016, Karki et al., 2020, Ding et al., 2021). A central mechanism for translational control is the phosphorylation of eIF2α, as even partial phosphorylation is sufficient to inhibit protein synthesis (Baird and Wek, 2012). Furthermore, rhythms in activity of the N. crassa eIF2α kinase CPC-3, a homolog of the well-studied yeast and mammalian eIF2α kinase GCN2, are dependent on rhythmic uncharged tRNAVal, levels. The rhythm in uncharged tRNAVal levels is driven, at least in part, by rhythms in valyl-tRNA synthetase (ValRS) levels (Karki et al., 2020). However, in addition to ValRS, several other aaRSs were found to be clock-controlled from genomic datasets (Sancar et al., 2015, Hurley et al., 2018, Castillo et al., 2022b), suggesting that aaRSs may be coordinately regulated by the clock to control rhythmic translation.
In this study, we sought to independently validate clock control of two aaRSs and begin to examine the mechanisms of clock control of aaRS genes. Using aspartyl-tRNA synthetase (AspRS) and glutaminyl-tRNA synthetase (GlnRS) luciferase translational reporters, we confirmed that AspRS and GlnRS protein levels are rhythmic in WT cells with a peak in the subjective night, similar to the peak time of ValRS (Karki et al., 2020), and arrhythmic in clock mutant Δfrq cells. We identified clock-controlled transcription factors that regulate AspRS::LUC and GlnRS::LUC rhythmic accumulation. We show that AspRS levels are low and arrhythmic in cells deleted for transcription factor NCU00275, and GlnRS levels are high and display a low amplitude rhythm in cells deleted for transcription factor ADV-1. These findings provide a basis for further studies investigating coordinate clock control of aaRSs via different clock-controlled transcription factors and the roles of rhythmic aaRS’s in rhythmic mRNA translation.
Strains, key reagents, and oligonucleotide primers used in this study are listed in Table 1. N. crassa wild type 74-OR23-IV (FGSC 4200) was grown in Vogel’s minimal media with 2% glucose (V2G) (Davis and De Serres, 1970). All strains containing the hygromycin phosphotransferase (hph) construct conferring resistance to hygromycin B were maintained on V2G and supplemented with 200 μg/mL of hygromycin B (VWR). Strains containing the bar cassette conferring resistance to Basta were maintained on V2G lacking NH4NO3 and supplemented with 0.5% proline (Sigma-Aldrich) and 200 μg/mL of Basta (LibertyTM, Bayer).
| Reagent or resource | Source | Identifier |
|---|---|---|
| Plasmids | ||
| Plasmid pRMP57 | Gooch et al., 2008 | GenBank KC890770.1 |
| Plasmid pBP15 | Beasley et al., 2006 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Basta (LibertyTM) | Bayer | Cat#280SL |
| Hygromycin | VWR | Cat#80055-268 |
| Luciferin | Gold Biotechnology | Cat#LUNCA-300 |
| Strains | ||
| Neurospora crassa wild type 74-OR23-IV mat a | FGSC | FGSC 4200; DBP 985 |
| Δfrq::bar, mat A | Bennett et al., 2013 | DBP 1228 |
| Δncu00275::hph, mat a | FGSC | FGSC 12217; DBP 927 |
| Δadv-1::hph, mat A | FGSC | FGSC 11042; DBP 917 |
| Δclr-1::hph, mat a | FGSC | FGSC 11029; DBP 981 |
| Δsah-1::hph, mat a | FGSC | FGSC 11132; DBP 990 |
| Δvos-1::hph, mat a | FGSC | FGSC 13536; DBP 1970 |
| WT, AspRS::LUC translational fusion | This paper | DBP 3999 |
| WT, GlnRS::LUC translational fusion | This paper | DBP 3991 |
| WT, GlnRS::LUC translational fusion | This paper | DBP 3992 |
| Δfrq::bar, AspRS::LUC translational fusion | This paper | DBP 4000 |
| Δfrq::bar, GlnRS::LUC translational fusion | This paper | DBP 3989 |
| Δncu00275:hph, AspRS::LUC translational fusion | This paper | DBP 4142 |
| Δclr-1::hph, AspRS::LUC translational fusion | This paper | DBP 4137 |
| Δncu00275::hph, GlnRS::LUC translational fusion | This paper | DBP 4211 |
| Δadv-1::hph, GlnRS::LUC translational fusion | This paper | DBP 4208 |
| WT, Pasprs::luc transcriptional fusion | This paper | DBP 4280 |
| WT, Pglnrs::luc transcriptional fusion | This paper | DBP 4283 |
| WT, Pasprs::luc transcriptional fusion | This paper | DBP 4416 |
| ∆frq::bar, Pasprs::luc transcriptional fusion | This paper | DBP 4419 |
| WT, Pglnrs::luc transcriptional fusion | This paper | DBP 4407 |
| ∆frq::bar, Pglnrs::luc transcriptional fusion | This paper | DBP 4411 |
| Oligonucleotides | ||
| To generate AspRS::LUC asprsF1 5′ CAAAGCAACATGGCCGACAG 3′ | This paper | N/A |
| To generate AspRS::LUC asprsR1 5′ TGGCGTCCTCAGGAAGCAACCTCTTGGGCG 3′ | This paper | N/A |
| To generate AspRS::LUC asprsF2 5′ GTTGCTTCCTGAGGACGCCAAGAACATCAA 3′ | This paper | N/A |
| To generate AspRS::LUC asprsR2 5′ ATGAAGTCACTTAATCAGACGGCGATCTTG 3′ | This paper | N/A |
| To generate AspRS::LUC asprsF3 5′ GTCTGATTAAGTGACTTCATTGTCGGTGGG 3′ | This paper | N/A |
| To generate AspRS::LUC asprsR3 5′ GATAACTGAAGGCTCGAAAT 3′ | This paper | N/A |
| Validation of endogenous integration of AspRS::LUC asprsF4 5′ GCGATGGCATGCTGCCGACG 3′ | This paper | N/A |
| Validation of endogenous integration of AspRS::LUC asprsR4 5′ GTCAGCTTGCTTCCCATAAG 3′ | This paper | N/A |
| To generate GlnRS::LUC glnrsF1 5′ CAACTGCTACCTCCGATTCG 3′ | This paper | N/A |
| To generate GlnRS::LUC glnrsR1 5′ TGGCGTCCTCGTTCTTCTCCTTATCCTCCT 3′ | This paper | N/A |
| To generate GlnRS::LUC glnrsF2 5′ GGAGAAGAACGAGGACGCCAAGAACATCAA 3′ | This paper | N/A |
| To generate GlnRS::LUC glnrsR2 5′ GGACAACTGCTTAATCAGACGGCGATCTTG 3′ | This paper | N/A |
| To generate GlnRS::LUC glnrsF3 5′ GTCTGATTAAGCAGTTGTCCGCGAATTAAC 3′ | This paper | N/A |
| To generate GlnRS::LUC glnrsR3 5′ TAGAGGTAGTACTGCCAGCG 3′ | This paper | N/A |
| Validation of endogenous integration of GlnRS::LUC glnrsF4 5′ TGAAGTTTGGTGATGTCTCC 3′ | This paper | N/A |
| Validation of endogenous integration of GlnRS::LUC glnrsR4 5′ TCCGAATAGTACTTCTGTGG 3′ | This paper | N/A |
| To generate Pasprs::luc asprsF5 5′ ATCGGCGGCCGCACGGATTATACGATGCCCGC 3′ | This paper | N/A |
| To generate Pasprs::luc asprsR5 5′ CGATACTAGTGTTGCTTTGTCGAATTCGAT 3′ | This paper | N/A |
| To generate Pglnrs::luc glnrsF5 5′ ATCGGCGGCCGCGTATTGAAAATAGGTGGGGA 3′ | This paper | N/A |
| To generate Pglnrs::luc glnrsR5 5′ CGATACTAGTGATGTGTCTGTGTGTGTGGT 3′ | This paper | N/A |
| Software and algorithms | ||
| BioDare2 (beta version 2) | Zielinski et al., 2014 | https://biodare2.ed.ac.uk/ |
| Cosine Wave Analysis | Lamb et al., 2011 | PRISM software package (GraphPad Software) |
| ECHO | De Los Santos et al., 2020 | https://github.com/delosh653/ECHO |
| ggplot2 R package | Wickham, 2016 | https://ggplot2.tidyverse.org |
| GraphPad Prism version 9.4.0 | https://www.graphpad.com/ | |
| Serial Cloner version 2.6.1 | http://serialbasics.free.fr/Serial_Cloner.html | |
| Other | ||
| EnVision Xcite Multilabel Reader | PerkinElmer | Cat#2105-0010 |
| NanoDropTM Microvolume Spectrophotometer | Thermo Scientific | Cat#ND-ONE-W |
To assay rhythmic translation of aspartyl tRNA synthetase (AspRS, NCU00915) and glutaminyl tRNA synthetase (GlnRS, NCU07926), an aaRS::LUC translational fusion to luciferase was generated by 3-way fusion polymerase chain reaction (PCR). Primers RSF1 and RSR1 were used to make fragment 1, and primers RSF3 and RSR3 were used to make fragment 3, both using wild type (WT) genomic DNA as template. Primers RSF2 and RSR2 were used to make fragment 2 using pRMP57, a plasmid containing the N. crassa codon-optimized luciferase gene as template DNA (Gooch et al., 2008). The three PCR fragments with overlapping regions were stitched via fusion PCR using primers RSF1 and RSR3, and the resulting PCR product was co-transformed with the plasmid pBP15 containing hph (Beasley et al., 2006) into WT (FGSC2489). Primers were designed using Serial Cloner version 2.6.1 and the sequences are given in Table 1. Lyophilized PCR oligonucleotide primers were obtained from Integrated DNA Technologies (IDT) and resuspended in 1x Tris-EDTA, pH 8.0 buffer to make 100 μM primer stocks. The PCR reaction mix was as follows: total reaction mix = 50 μl, water = 37.5 μl, 5X High Fidelity (HF) buffer = 5 μl, dNTP mix = 1 μl (10 mM), primers = 2.5 μl each (10 μM), Phusion® DNA polymerase = 0.5 μl (1.0 units/50 μl PCR), and template DNA = 1 μl (50 ng). Phusion® High Fidelity DNA polymerase kit (Cat. No. M0530) and dNTP mix (Cat. No. 4030) were purchased from New England Biolabs (NEB) and Takara, respectively. The following annealing temperatures and extension times were applied: AspRS fragment 1 = 65°C, 1:30 min, fragment 2 = 72oC, 1:30 min, fragment 3 = 59°C, 1:30 min, and fragments 1+2+3 = 59°C, 3 min; GlnRS fragment 1 = 63°C, 1:30 min, fragment 2 = 72°C, 1:30 min, fragment 3 = 64oC, 30 s, and fragments 1+2+3 = 63°C, 3 min. PCR cycling was performed with a C1000 Touch Thermal Cycler (Bio-Rad) using the following program: 30 cycles of denaturation at 98°C for 10 s, annealing at varying temperatures for 30 s (as described above) and extension at 72°C for varying times (as described above).
Hygromycin-resistant transformants were screened for luciferase activity and homologous insertion into the aars gene (primers RSF4 and RSR4) using the same PCR conditions described above with annealing temperatures and extension times for AspRS = 62°C, 4 min, and GlnRS = 59°C, 4 min. To generate aaRS::LUC in different mutant strains, aaRS::LUC, WT were crossed with the knockouts on synthetic cross medium supplemented with 0.25% biotin (Westergaard and Mitchell, 1947, Davis and De Serres, 1970). AspRS::LUC transformants were crossed with Δfrq::bar (DBP 1228) to generate AspRS::LUC, WT (DBP 3999), AspRS::LUC, Δfrq::bar (DBP 4000). GlnRS::LUC transformants were crossed with Δfrq::bar (DBP 1228) (Bennett et al., 2013) to generate GlnRS::LUC, WT (DBP 3991 mat A and DBP 3992 mat a), GlnRS::LUC, Δfrq::bar (DBP 3989). DBP 3999 was crossed with Δclr-1::hph (DBP 981) and Δncu00275 (DBP 927) to generate AspRS::LUC, Δclr-1::hph (DBP 4137) and AspRS::LUC, Δncu00275::hph (DBP 4142), respectively. DBP 3992 was crossed with Δadv-1::hph (DBP 917) to generate GlnRS::LUC, Δadv-1::hph (DBP 4137). DBP 3991 was crossed with Δncu00275::hph (DBP 927) to generate GlnRS::LUC, Δncu00275::hph (DBP 4211).
To generate transcriptional fusions to luc, promoter regions of asprs (primers asprsF5 and asprsR5 to generate the 1.8 kb fragment Pasprs), and glnrs (primers glnrsF5 and glnrsR5 to generate the 1.82 kb fragment Pglnrs) were amplified by PCR using the cycling conditions described above with annealing temperatures and extension times for fragments Pasprs and Pglnrs = 68oC, 1:30 min. PCR products were digested with NotI and SpeI (NEB), and cloned into plasmid pRMP57 containing the codon-optimized luciferase gene. The resulting plasmids were linearized by PciI (NEB) digest, co-transformed with hygR pBP15 into WT (FGSC 4200) cells, and hygromycin-resistant transformants were screened for luciferase activity. Pasprs::luc transformants were crossed with Δfrq::bar (DBP 1228) (Bennett et al., 2013) to generate Pasprs::luc, WT (DBP 4416) and Pasprs::luc, Δfrq::bar (DBP 4419). Pglnrs::luc transformants were crossed with Δfrq::bar (DBP 1228) to generate Pglnrs::luc, WT (DBP 4407), and Pglnrs::luc, Δfrq::bar (DBP 4411).
To examine bioluminescence rhythms arising from strains containing luciferase fusions, 5 μl of a 1×105 conidia/ml suspension were inoculated into 96 well microtiter plates containing 150 μl of 1X Vogel’s salts, 0.01% glucose, 0.03% arginine, 0.1 M quinic acid, 1.5% agar, and 25 μM firefly luciferin, pH 6. After inoculation of conidia (1×105 conidia), the microtiter plate was incubated at 30°C in LL for 24 h and transferred to DD 25°C to obtain bioluminescence recordings using EnVision Xcite Multilabel Reader (Perkin-Elmer), with recordings taken every 90 min over 4–5 days. Raw reads were normalized to the mean to graph the data.
Rhythmic expression of N. crassa clock-controlled tRNA synthetases was determined using public ribosome profiling (ribo-seq) and RNA-seq datasets for WT and clock mutant Δfrq cells (Castillo et al., 2022b). Briefly, ribo-seq and RNA-seq were performed in parallel from cells grown in constant darkness (DD) in a circadian time course with 4-h resolution for two biological replicates.
Rhythmic data from luciferase assays were fit to a sine wave or a line as previously described (Lamb et al., 2011). Nonlinear regression to fit the rhythmic data to a sine wave (fitting period, phase, and amplitude) and a line (fitting slope and intercept), as well as Akaike’s information criteria tests to compare the fit of each data set to the 2 equations, were carried out using the Prism software package version 9.4.0. The p-values reflect the probability that, for instance, the sine wave fits the data better than a straight line. Error bars in all graphs represent the standard error of the mean (SEM) from independent experiments. Raw and normalized luciferase activity reads were analyzed for period, phase, and amplitude values using BioDare version 2 (Zielinski et al., 2014). Heat maps were generated using the ggplot2 R package for genes with rhythmic ribosome protected fragments (RPF) counts in WT, and sorted according to increasing peak phase of the oscillation (Wickham, 2016). ribosome protected fragments (RPF) levels are standardized within each gene (row) (Z-scores).
Published N. crassa circadian ribosome profiling (ribo-seq) data revealed rhythms in ribosome occupancy for 17 of 36 aaRS using the Extended Circadian Harmonic Oscillator (ECHO) rhythmicity detection tool (De Los Santos et al., 2020, Castillo et al., 2022b). Genes with an adjusted p-value of < 0.05, and with circadian harmonic, damped, or forced oscillation types were considered rhythmic. A heat map of the phase-sorted fitted ribosome protected footprint (RPF) values obtained using ECHO showed robust rhythmic ribosome occupancy for 17 aaRSs in WT cells, with peak ribosome occupancy primarily during the late subjective day (DD40-44) (Figure 1A, Class I). As expected for circadian clock control, the rhythms were abolished in the clock mutant Δfrq cells. Class II aaRS genes were arrhythmic in both WT and Δfrq cells (Figure 1A). Class III aaRS genes were rhythmic in both WT and Δfrq cells, suggesting that the rhythms are controlled by something other than the FRQ/WCC circadian oscillator (Figure 1A). Circadian rhythms in ValRS protein levels were previously validated using a luciferase (LUC) translational reporter. ValRS::LUC levels peaked in the subjective night (Karki et al., 2020), lagging the observed peak in ribosome occupancy (Figure 1A).
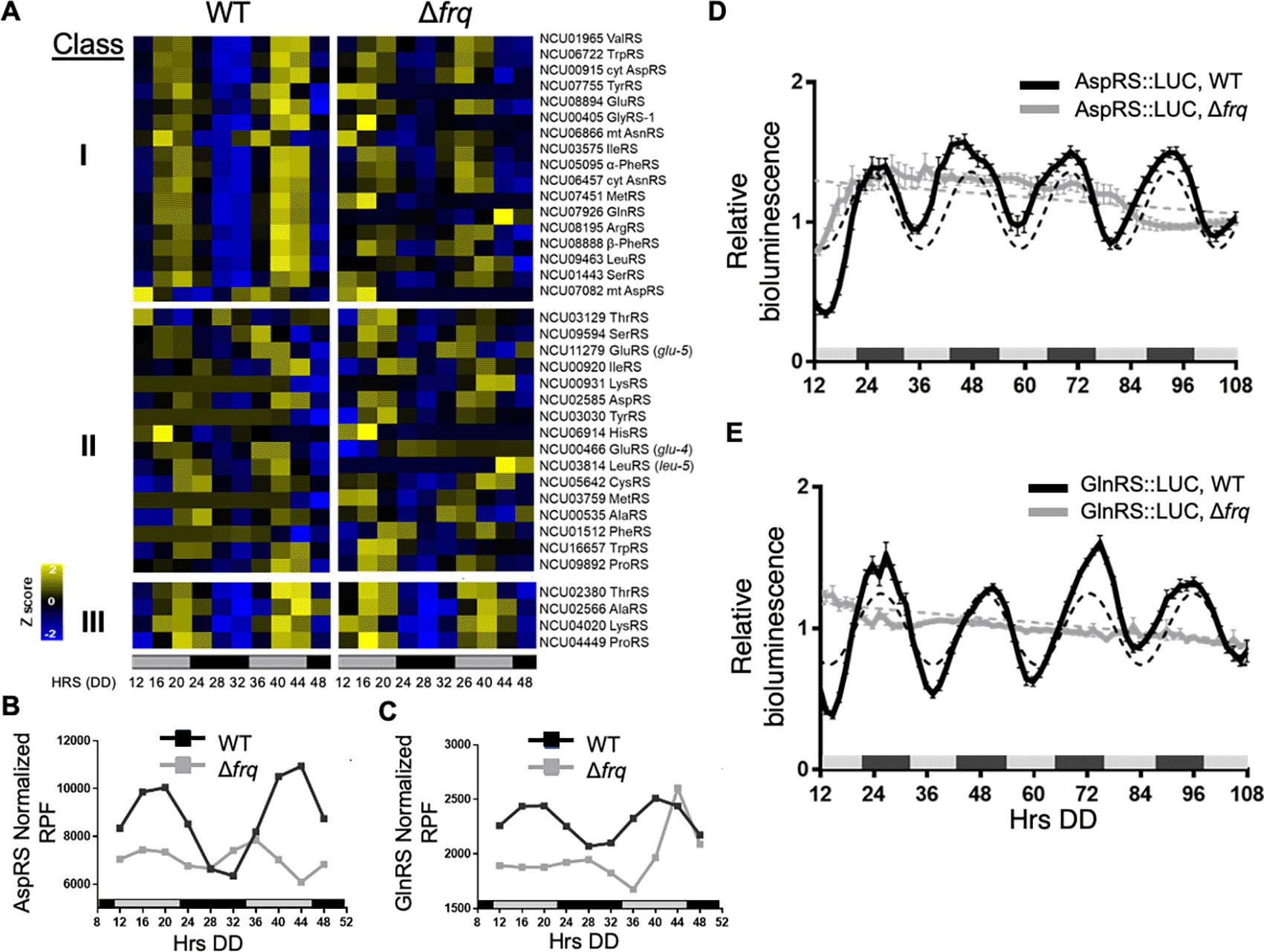
A) Heat maps of the peak phase of aaRS mRNAs with rhythmic ribosome protected fragments (RPF) counts in WT, and arrhythmic RPF counts in Δfrq cells (Class I), arrhythmic in both WT and Δfrq cells (Class II), or rhythmic in both WT and Δfrq cells (Class III) from two replicate samples grown in DD and harvested at the indicated times (Hrs). Genes in Class I are sorted by the peak phase in WT. Cyt (cytoplasmic), mt (mitochondrial) B-C. Plots show the normalized fitted RPF reads (n=2) in WT (black line, ECHO p-value ≤ 0.05) and Δfrq (gray line, ECHO p-value > 0.05 for AspRS and p-value = 0.04 for GlnRS with a short 16 h period) cells for B) AspRS and C) GlnRS. Luciferase (LUC) activity from D) AspRS::LUC and E) GlnRS::LUC translational fusions in WT (black line) and Δfrq (gray line) cells. The average bioluminescence signal is plotted (AspRS::LUC, mean ± SEM, n=12 and GlnRS::LUC, mean ± SEM, n=24). Raw reads were normalized to the mean to plot the data. AspRS::LUC and GlnRS::LUC in WT cells were rhythmic as indicated by a better fit to a sine wave (dotted black line, p-value < 0.001). AspRS::LUC and GlnRS::LUC in Δfrq were arrhythmic as indicated by a better fit of the data to a line (dotted gray line p-value > 0.05). The bar at the bottom of the heat maps and graphs represents subjective day (gray) and subjective night (black) in this and all subsequent figures. Data for A-C from Castillo et al. (2022b).
In higher eukaryotes, 9 aaRSs form a multisynthetase complex (MSC) that is proposed to aid translation by providing a channel through which tRNAs can pass to reach bound aaRSs (Hyeon et al., 2019). Interestingly, 5 of the 9 aaRSs in the complex (AspRS, GlnRS, GluRS, LeuRS, and MetRS) are clock-controlled based on our ribosome profiling datasets (Figure 1A), and we focused on validating circadian clock control of AspRS and GlnRS (Figure 1B & C). AspRS and GlnRS luciferase translational reporter fusions were generated (AspRS::LUC and GlnRS::LUC) and assayed for rhythmic luciferase levels from cells grown in DD over 4 days (Figure 1D & C). Bioluminescence rhythms were observed for both AspRS::LUC and GlnRS::LUC, with peak levels during the early subjective night (e.g. DD 48) and a period of 22.4 ± 0.6 h and 22.0 ± 0.4 h, respectively (Table 2). Similar to ValRS::LUC, the peak in AspRS::LUC and GlnRS::LUC levels occurred a few hours after the peak in ribosome occupancy (Figure 1B-E). The AspRS::LUC and GlnRS::LUC rhythms were abolished in Δfrq cells, confirming that AspRS and GlnRS protein levels are clock-controlled (Figure 1D & E, Table 2).
In addition to rhythms in protein levels, several N. crassa aaRS mRNAs were reported in genome-wide studies to be clock-controlled (Hurley et al., 2014, 2018, Sancar et al., 2015, Castillo et al., 2022b). Of these aaRSs, asprs and glnrs exhibited rhythms in mRNA levels in WT cells, with mRNA levels peaking in the subjective early evening (DD40-44) (Figure 2A & B). In support of these genomic data, asprs (Pasprs::luc) and glnrs (Pglnrs::luc) promoter luc fusions were rhythmic in DD peaking during the subjective night (Figure 2C & D), with no significant period and phase differences between the mRNA and protein levels (Table 2).
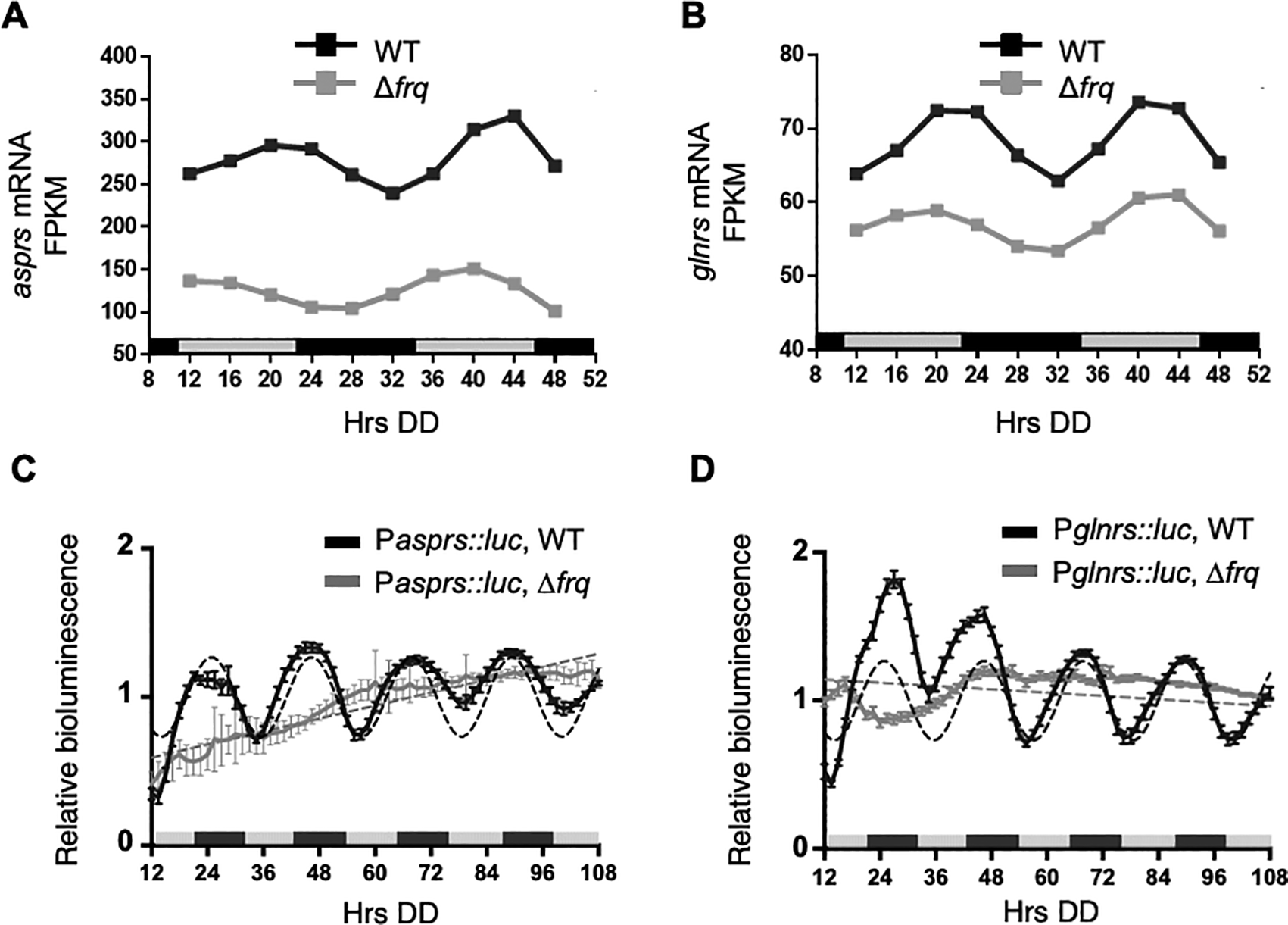
A-B. Plots show the mRNA fragments per kilobase of exon per million mapped fragments (FPKM) levels in WT (black line, ECHO p-value < 0.05) and Δfrq (gray line, ECHO p-value > 0.05) cells for A) asprs and B) glnrs. C-D. Plots show the luciferase activity from C) Pasprs::luc transcriptional (black line) and D) Pglnrs::luc transcriptional (black line) fusions in (black line) and ∆frq (grey line) cells grown in DD and recorded every 90 min over 4 d (Hrs DD). The average bioluminescence signal is plotted (mean ± SEM, n=12). Luciferase activities are rhythmic as indicated by a better fit to a sine wave (dotted black line, p-value < 0.0001) and arrhythmic in ∆frq cells where the best fit is to a line (dotted grey line). Data for A-B from Castillo et al. (2022b).
As expected for clock control, the mRNA rhythms were abolished in Δfrq cells as shown by the ECHO-generated fitted values for normalized mRNA levels by FPKM (fragments per kilobase of exon per million mapped reads) and by aars promoter luciferase reporter fusion assays (Figure 2A & B) (Castillo et al., 2022b). Together, these data support that rhythmic AspRS and GlnRS protein levels arise, at least in part, from clock-controlled mRNA levels.
To determine if clock-controlled rhythms in AspRS and GlnRS protein levels require clock-controlled transcription factors and rhythmic transcription, we examined AspRS::LUC and GlnRS::LUC rhythms in cells deleted for transcription factors that are direct targets of the WCC. The levels of the transcription factor mRNAs peak in the subjective morning and the transcription factors bind to downstream ccgs to regulate their rhythmic expression (Smith et al., 2010, Dekhang et al., 2017, Munoz-Guzman et al., 2021). Three clock-controlled transcription factors were selected for this initial study. CLR-1 (NCU07705) is a zinc binuclear cluster transcription factor that is important in nutrient sensing and signaling, and was shown to be necessary for normal AspRS mRNA levels when N. crassa cells were grown on cellulose (Coradetti et al., 2012). However, AspRS::LUC levels were rhythmic in Δclr-1 cells with a period and phase, and overall levels, that were similar to WT when the cultures were grown in DD (Figure 3A, Table 2). Transcription factor NCU00275 was selected as a representative clock-controlled transcription factor of unknown function (Smith et al., 2010). AspRS::LUC rhythms were abolished in Δncu00275 cells (Figure 3B, Table 2) and AspRS::LUC levels were lower in Δncu00275 compared to WT cells (Figure 3C). These data supported that NCU00275 is necessary for circadian rhythms in AspRS::LUC levels and directly, or indirectly, activates asprs transcription. GlnRS::LUC was rhythmic in Δncu00275 cells with no significant differences in levels, period or phase between WT and Δncu00275 (Figure 4A, Table 2). Previous RNA-seq data comparing WT to cells deleted for the transcription factor ADV-1 (NCU07392) showed that the levels of glnrs mRNA, but not asprs mRNA levels, were generally higher in Δadv-1 cells compared to WT grown in DD and following light treatments (Dekhang et al., 2017). ADV-1 ChIP-seq data did not reveal significant binding of ADV-1 to the promoter of glnrs, suggesting indirect negative regulation of glnrs by ADV-1. Based on these data, we examined GlnRS::LUC rhythms in Δadv-1, and found that GlnRS::LUC rhythms became progressively damped by day 3 in DD compared to WT (Figure 4B), leading to a significantly reduced amplitude of oscillation (Figure 4C, Table 2). Consistent with the RNA-seq data, the raw bioluminescence signals showed that GlnRS::LUC levels were higher in Δadv-1 than in WT cells (Figure 4D). Taken together, these data support that specific clock-controlled transcription factors contribute to the rhythmic expression of aaRSs.
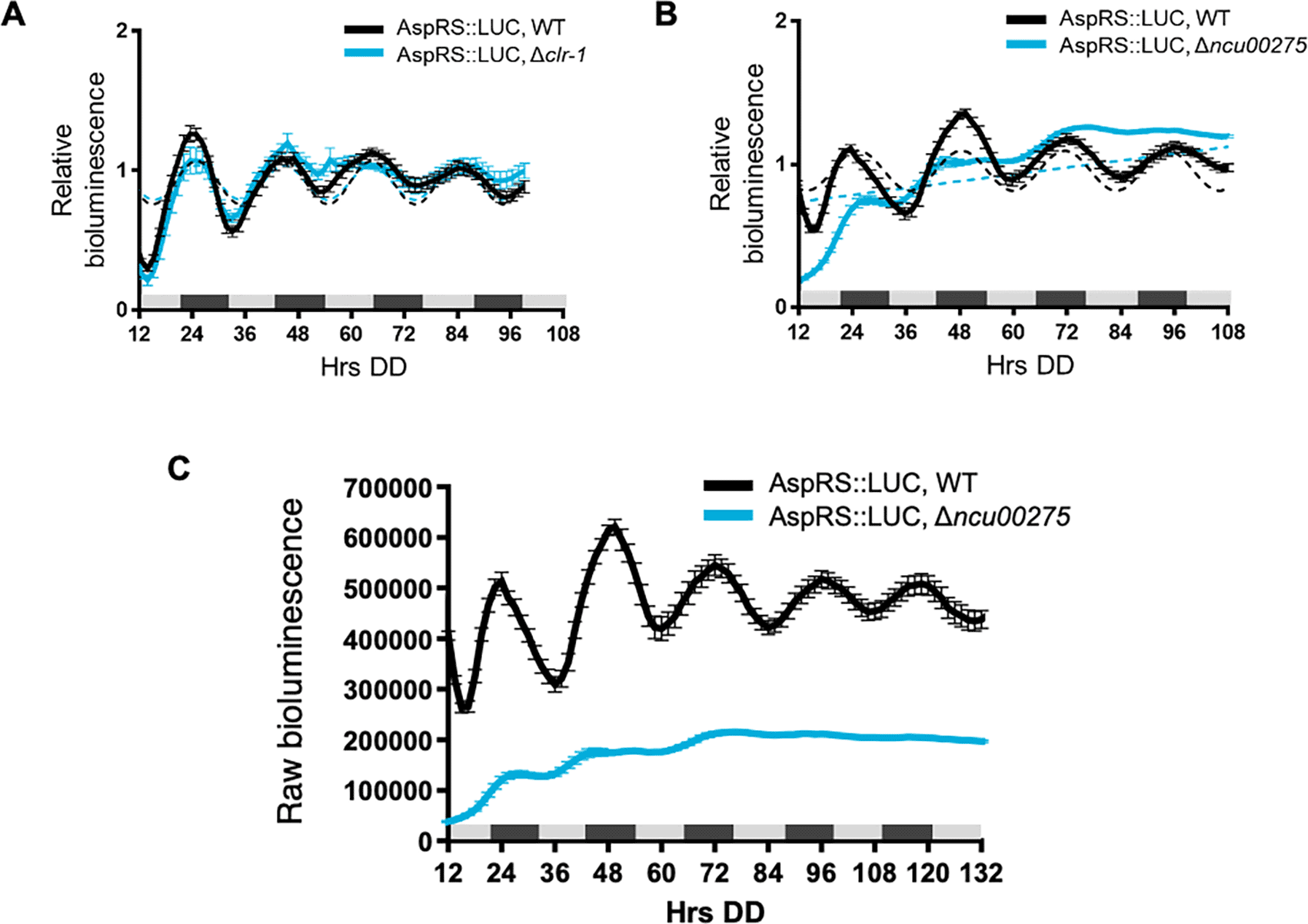
Luciferase activity from AspRS::LUC translational fusions in WT (black line) and transcription factor knockouts A) Δclr-1 (blue line) and B) Δncu00275 (blue line). Raw reads were normalized to the mean to plot the data. The average bioluminescence signal is plotted (mean ± SEM, n=12). Luciferase activities are rhythmic as indicated by a better fit to a sine wave (dotted black line, p-value < 0.0001) or arrhythmic as indicated by a better fit of the data to a line (dotted blue line, p-value > 0.0001). C) Raw bioluminescence signals from AspRS::LUC translational fusions in WT (black line) and transcription factor knockout Δncu00275 (blue line).
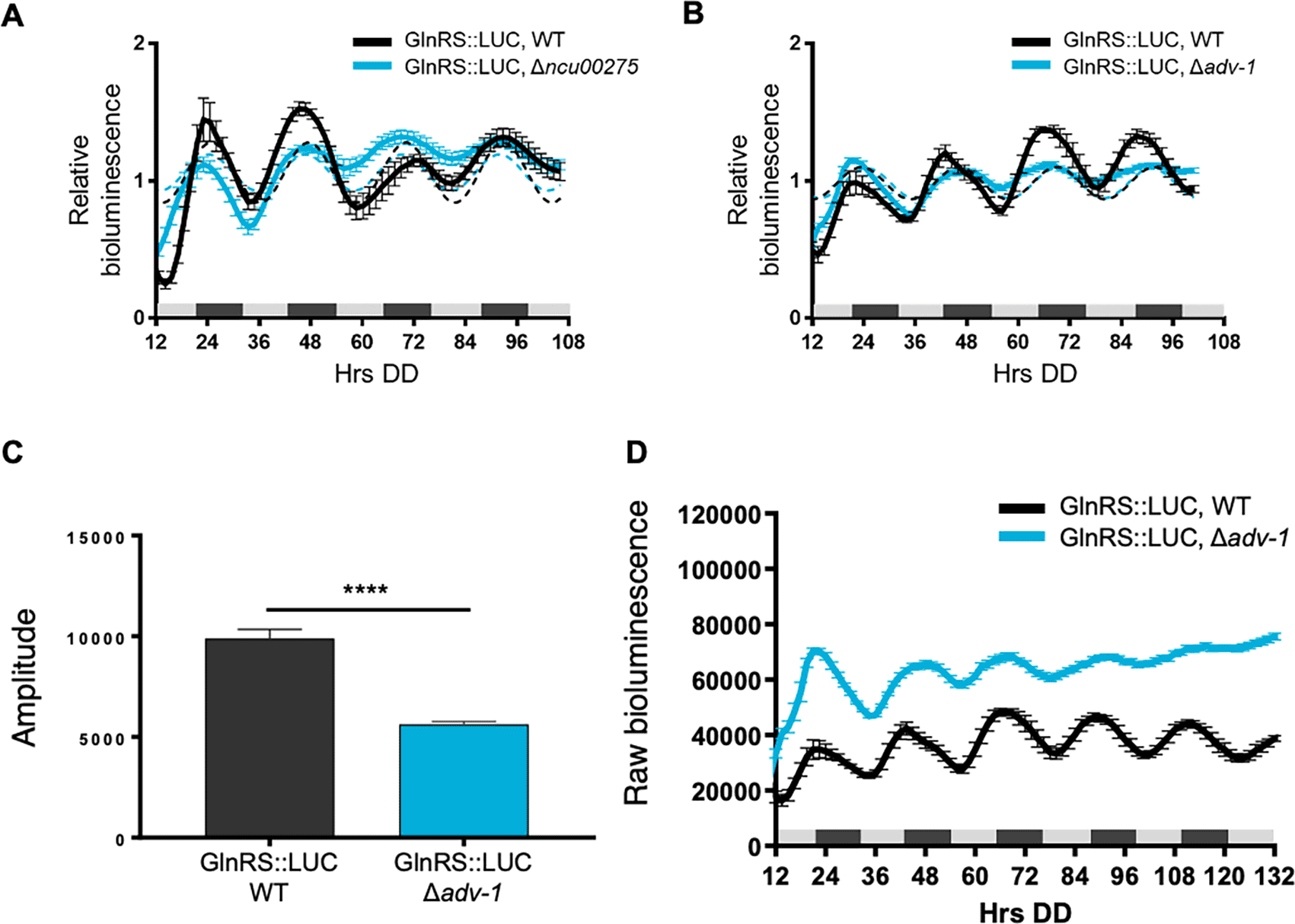
Luciferase activities from GlnRS::LUC translational fusions in WT (black line) and transcription factor knockouts A) Δncu00275 (blue line) and B) Δadv-1 (blue line). The average bioluminescence signal is plotted (mean±SEM, n=12). Luciferase activities are rhythmic as indicated by a better fit to a sine wave (dotted black line, p-value<0.0001) or arrhythmic as indicated by a better fit of the data to a line (dotted blue line, p-value>0.0001). C) Mean amplitude (mean ± SEM, n=12; ****p-value < 0.0001) of GlnRS::LUC bioluminescence traces in WT (black bar) and Δadv-1 (blue bar) cells. P-values were calculated by an unpaired t-test. D) Raw bioluminescence signals from GlnRS::LUC translational fusions in WT (black line) and transcription factor knockout Δadv-1 (blue line). The average bioluminescence signal is plotted (mean±SEM, n=12).
All underlying data can be found in the Underlying data section (Castillo et al., 2022b, Castillo and Bell-Pedersen, 2022).
aaRSs play a central role in translation and translation fidelity, yet the regulation of aaRS gene expression in eukaryotes is understudied. Genome-wide datasets revealed that several eukaryotic aaRSs are clock-regulated at the level of mRNA and protein (Sancar et al., 2015, Hurley et al., 2018, Castillo et al., 2022b, Barclay et al., 2012, Pembroke et al., 2015, Hughes et al., 2009, Miller et al., 2007, Vollmers et al., 2012, Eckel-Mahan and Sassone-Corsi, 2009, Geyfman et al., 2012, Yoshitane et al., 2014, Janich et al., 2015). We previously showed that the levels of ValRS cycle under control of the circadian clock with peak levels during the subjective night (Karki et al., 2020). Here, we validated that asprs and glnrs mRNA and protein levels are also clock-controlled with a similar night-time peak in protein levels. The bulk of rhythmic protein accumulation occurs at night in N. crassa (Hurley et al., 2018) supporting that the night-time peak in aaRS protein levels serve to coordinately increase protein synthesis at night.
We observed that AspRS and GlnRS protein rhythms are dependent on the circadian clock through the activities of clock-controlled transcription factors. AspRS levels are arrhythmic and low in Δncu00275, suggesting that NCU00275 activates asprs transcription. NCU00275 is annotated as a hypothetical protein, but its homologs in other fungi suggest that it is a C3HC4-type RING finger protein involved in transcription, signal transduction, ubiquitination, and recombination (Basenko et al., 2018, Krishna et al., 2003). NCU00275 mRNA levels peak in the subjective morning under control of the WCC (Smith et al., 2010), whereas asprs mRNA and protein levels peak several hours later in the early subjective night. This delay suggests the possibility that the effect of NCU00275 on asprs transcription is indirect, requires other transcription factors or changes in chromatin state, and/or that posttranscriptional regulation of asprs contributes to this delay. Furthermore, deletion of NCU00275 had no significant effect on GlnRS levels, indicating that coordinate regulation of the aaRS’s by the clock is not necessarily through the same clock-controlled transcription factors. Consistent with this idea, the clock-controlled transcription factor ADV-1 was previously shown to affect the levels of glnrs, but not asprs, mRNA levels (Dekhang et al., 2017), and GlnRS levels and rhythms were altered in Δadv-1 cells compared to WT. Furthermore, ChIP-seq data showed that ADV-1 binds to the promoter of TrpRS (NCU06722) and MetRS (NCU07451), both of which are clock-controlled, and to the promoter of LysRS (Dekhang et al., 2017), which was not rhythmic in our ribo-seq data (Figure 1). Additional experiments will be done in the future to confirm clock control of the other N. crassa aaRS’s predicted to be rhythmic from RNA-seq and ribo-seq datasets, and to comprehensively define the roles of clock-controlled transcription factors in aaRS regulation.
Charging tRNAs with the correct amino acid is the first step in translation, and therefore the levels and function of aaRSs are critical to translation fidelity (Yu et al., 2021, Hausmann and Ibba, 2008). Mistakes in translation are generally considered detrimental; however, during stress, mistranslation may be beneficial by increasing the levels of altered proteins that can perform new functions to aid the response (Pan, 2013, Ribas de Pouplana et al., 2014). In eukaryotes, some aaRSs form the MSC (Bandyopadhyay and Deutscher, 1971, Lee et al., 2004, Kerjan et al., 1994), with varying composition dependent on the organism. In S. cerevisiae, the MSC is comprised of MetRS, GluRS and the scaffold protein Arc1 (Galani et al., 2001). The MSC in mammals has 9 aaRSs, including 5 of the 9 aaRSs (AspRS, GlnRS, GluRS, LeuRS, and MetRS) that are rhythmic in ribo-seq data sets, and 3 scaffold proteins, AIMP1-3. The MSC helps the function of its components; for example, the Km for binding of tRNAMet to MetRS in the yeast MSC is about 100-fold lower compared to the Km for binding of tRNAMet to MetRS alone (Simos et al., 1998). MSC components are also involved in cell signaling, stress responses, metabolite sensing, and controlling gene expression by binding to specific RNA and DNA sites, supporting the idea that a key role of the MSC is to support alternative functions of aaRSs (Cui et al., 2021, Pang et al., 2014, Rubio Gomez and Ibba, 2020). Furthermore, aaRSs, either alone or in the MSC complex, participate in a wide variety of processes outside of their classical role in tRNA charging, including transcription regulation, splicing, and metabolism (Rubio Gomez and Ibba, 2020), and abnormal expression, localization, and molecular interactions of aaRSs are associated with a variety of human diseases, including cancer (Zhou et al., 2020). This widespread impact of aaRSs on host biology raises the intriguing idea that daily rhythms in the levels of aaRSs represent a missing factor linking the clock to a wide range of rhythmic biological processes that are critical to health, underscoring the need to better understand the mechanisms underlying their circadian regulation.
Gene Expression Omnibus: Ribosome profiling and RNA-seq data used in Figures 1 and 2. Accession number GSE181566; https://identifiers.org/geo:GSE181566 (Castillo et al., 2022b).
Figshare: Circadian Clock Control of tRNA synthetases in Neurospora crassa. https://doi.org/10.6084/m9.figshare.c.6209830.v4 (Castillo and Bell-Pedersen, 2022).
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Circadian clocks Neurospora biology
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: circadian rhythm, translational control, neuroscience
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Circadian rhythms, Neurospora genetics and gene expression
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: circadian rhythm, translational control, neuroscience
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 17 Aug 23 |
read | read |
|
Version 1 22 Dec 22 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)