Keywords
proteomics, network analysis, Cytoscape app, Cytoscape, mechanistic hypotheses, curated signaling
This article is included in the Cytoscape gateway.
This article is included in the Bioinformatics gateway.
This article is included in the Cell & Molecular Biology gateway.
proteomics, network analysis, Cytoscape app, Cytoscape, mechanistic hypotheses, curated signaling
This version changes both to this manuscript and the Cytoscape app in response to reviewer comments. Changes focused on clarifying the Methods section to simplify the app's usage. Additionally, we have fixed a bug within the project code identified by the reviewer, and users can now create multiple networks with the tool. Lastly, in Figure 1, we have added a new sub-panel to highlight additional examples.
See the authors' detailed response to the review by Ruth Isserlin
CausalPath is a recently developed software tool that allows users to map proteomic and other molecular profiles to pathway information from the Pathway Commons database (RRID: SCR_001749) and other resources.1–4 CausalPath focuses on providing users a means to assess causality among correlated measurements. CausalPath can generate a network model of molecular signal flow that is consistent with both the profiling data and the published literature (Figure 1A). Results in the generated network model are linked to Pathway Commons, an integrative database of molecular interactions, and literature connections, therein.3,5
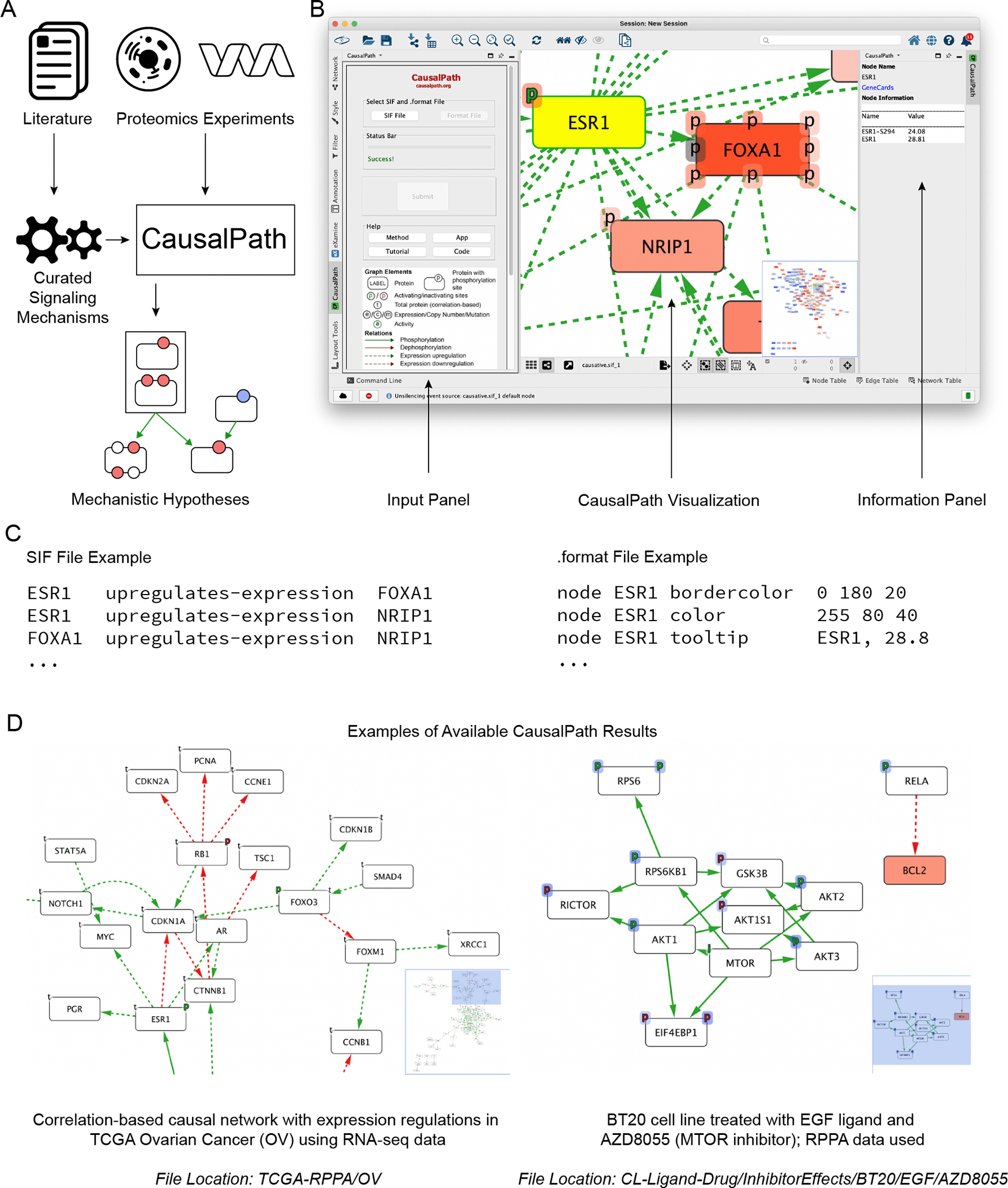
A: CausalPath maps data from experiments and curated literature resources to provide users with mechanistic hypotheses of experimental observations. B: CausalPath results for luminal (luminal A and luminal B) samples compared to basal-like samples; samples were profiled via proteomic mass spectrometry. The right-hand panel shows differential abundance measures for the ESR1 along with measured phosphorylation sites (e.g., ESR1 S294). Note: ESR1 appears yellow because Cytoscape uses yellow for network selections; unselected ESR1 would appear red, in this example, because ESR1 has an increased abundance in luminal versus basal breast cancer. C: Snippets of the SIF and .format input files are shown. D: Two sample CausalPath network results from those available in the Zenodo archive provided in the data availability section. Note: The causative .sif and .format files were loaded. Left: Correlation-based causal network with expression regulations in TCGA Ovarian Cancer (OV) using RNA-seq data; File Location: TCGA-RPPA/OV; Right: BT20 cell line treated with EGF ligand and AZD8055 (MTOR inhibitor); RPPA data used; File Location: CL-Ligand-Drug/InhibitorEffects/BT20/EGF/AZD8055.
Several tools currently support the visualization of CausalPath, including causalpath.org, Newt, and ChiBE.2,6,7 A comparison of unique features of these tools with respect to usage with CausalPath has been previously discussed in one of our recent publications.1 The CausalPath Cytoscape app provides a tool for importing, visualization, and utilizing CausalPath results from within Cytoscape, thereby facilitating user access to additional analyses not found in the aforementioned CausalPath tools.8 Cytoscape is a widely used extensible bioinformatics environment for the analysis and visualization of biological networks with nearly 10 million downloads since 2014.8 By providing users with a Cytoscape-based interface for the visualization of CausalPath results, users have access to the Cytoscape ecosystem of functionality. Briefly, this allows CausalPath users to have access to Cytoscape features such as network layouts and sharing of CausalPath results through to Network Data Exchange (NDEx).9 Additionally, various secondary analyses are available for users to run including, for example, gene set analysis through tools (e.g., through NDEx Integrated Query), module detection, and various network propagation methods.8
The work is implemented in the Java 16 programming language using the Cytoscape software platform. Multiple Cytoscape application programming interfaces (APIs) have been used to call the different functionalities. The CausalPath Cytoscape App13 is described in the three following sections:
The minimum system requirements for use of the CausalPath Cytoscape app include: Hardware: CPU: 1 GHz CPU, Memory: 512MB, Monitor: 1024×768 resolution; Software: Java 11.
To install the CausalPath Cytoscape app users must first install. Next, users must install the enhanceGraphics app from the “Apps -> App Manager …” menu of Cytoscape. After this, users can then install the CausalPath Cytoscape app App Manager menu.
A collection of results from previous work is available on Zenodo12 and will be used here as input files for the CausalPath Cytoscape app; users should download this archive as part of this initial attempts to try out the app.
Users are strongly advised to read the two previous CausalPath publications.1,2 These will help the reader understand the CausalPath algorithm and learn how to run a CausalPath analysis; this must be done separately from Cytoscape. CausalPath analysis can be conducted online via the CausalPath site and we have recently published a detailed protocol for how users can do this locally on their computers.1 The protocol provides guidance on available options for conducting a CausalPath analysis.
Once installed (see Installation section) users can visualize the results using the CausalPath Cytoscape app and run any additional analyses available from Cytoscape. The steps to visualize CausalPath results in Cytoscape are as follows. First, with Cytoscape open, users enable the CausalPath by clicking the App menu and then CausalPath. Next, from the Cytoscape Control Panel menu that appears, users will load both their SIF network and .format file. Lastly, users need to click the “Submit” button to visualize the network.
The user interface consists of two panels (as seen in Figure 1):
1 Input Panel: A panel component that appears on the left that is built using Cytoscape swing-application-api where users select files from their local file storage. The Java Swing library is used to choose the appropriate file and the Cytoscape io-api to read the network file. The panel contains a progress bar to show the progress and a help button that uses Cytoscape service-api to redirect to the CausalPath project website.
2 Information Panel: A panel component appearing on the right where a node or edge-specific information is shown when a user selects a node or edge.
Data from the network and format files is stored using multiple data structures (e.g., HashMaps with different class types and Arrays). A network view is created using Cytoscape view-model-API for the network and different styles for each node and edge are configured using Cytoscape presentation-api. Finally, we used the Cytoscape work-api to show the network. By default, the app lays out the network using a force-directed layout.
CausalPath visualizations include annotations on nodes (i.e., small circles attached to nodes). These annotations provide information on post-translational modifications (i.e., phosphorylations) as well as information on non-proteomic data (e.g., gene expression data and copy number information). To provide these annotations, we have made use of the Cytoscape enhancedGraphics plugin and the vizmap-api to map the sites to each node and we have used the “Label” chart type. Annotations are placed on the border of nodes when they are present in the input data (e.g., the example in Figure 1). This app uses the Cytoscape vizmap-api to map the sites to each node and we have used the “Label” charts style from the enhanced Graphics plugin to design the site annotations for each node. The visual notation of CausalPath uses a notation outlined previously1,2 with one exception. Instead of using border colors to indicate the activatory or inhibitory nature of the site, we used the text color of the letter within the annotation circle. This change was done to Cytoscape as this was the representation available with vizmap-api labels chart.
The CausalPath Cytoscape App requires two input files to visualize CausalPath result networks:
• Network file (with a .sif extension): The network data contains multiple rows where each row consists of tab-separated values where the first value refers to the source node, the second value refers to the edge type, and the third one refers to the target node.
• Format File (with a .format extension): The.format file also has multiple rows where each row consists of space and pipe-separated values. Each row can contain one of the multiple possible categories of information. The categories include: 1) styling for a node (e.g. color, border color, border width), 2) styling information for each site-related annotation linked to a particular node, or 3) “tooltip” information that will appear in the right-hand information panel, such as specific measured node values. More details can be found on this GitHub repository.
Figure 1 (partial view of the network) is a visualization of the results generated by CausalPath for a dataset taken from the National Cancer Institute's Clinical Proteomic Tumor Analysis Consortium (CPTAC) (RRID:SCR_017135) breast cancer study that collected proteomic and phosphoproteomic profiles for 105 of the original The Cancer Genome Atlas (TCGA) (RRID:SCR_003193) breast cancer samples with mass spectrometry; see data availability section for information on the input files used in this section.10 Specifically, Figure 1 shows luminal (luminal A and luminal B) samples compared to basal-like samples. Briefly, these results highlight overexpression of the ESR1 gene and genes directly downstream of ESR1 in luminal samples as compared to basal-like samples; a key characteristic of basal-like breast cancer is that it is hormone-receptor negative (estrogen-receptor and progesterone-receptor negative).11
The causative.sif and causative.format input files (Figure 1C) from the LumAB-vs-Basal folder are loaded by using the clicking app “SIF File” and “Format File” buttons of the Input Panel shown in Figure 1B; the full path to these two input files within the Zenodo12 archive is CausalPath-data/CPTAC-BRCA/subtypes/LumAB-vs-Basal; see data availability section for information on the Zenodo archive and the data formats section for a description of the contents of .sif and .format files. After clicking the “Submit” button users will see the resulting output visualization as shown in Figure 1B. For node or edge-specific information, the user should select that node or edge. The information regarding that node or edge will be shown in the side panel as shown in Figure 1. Users can also visualize multiple networks at the same time by repeating the instructions to load networks in the “Installation and usage” section. Additionally, we note for readers that the Zenodo archive provided in the data availability section12 provides pre-computed results for many of the cancer types (e.g., ovarian, bladder, renal, thyroid, pancreatic, colon, etc.) for which there is TCGA proteomics data. These CausalPath results for each of these cancer types can be visualized in the same manner as described above.
The CausalPath Cytoscape app is a software tool for visualizing network models with consistency with both the profiling data and the published literature. Use of CausalPath aids researchers who seek to understand the results of their experimental data and who would otherwise manually explore scientific literature to assess concordance with existing findings. By providing this tool as a Cytoscape app, we simplify the installation and use of CausalPath with existing analysis pipelines that combine other analyses available through Cytoscape that, for example, include a gene set and network modularity analyses.
The source data used in Figure 1 comes from a previous study using CausalPath results for LuminalAB-vs-Basal-like breast cancer comparison.2 The CausalPath input data and analysis results are publicly available on Zenodo (https://doi.org/10.5281/zenodo.4477801).12
The CausalPath Cytoscape app is available from the Cytoscape App Store: https://apps.cytoscape.org/apps/causalpathcytoscapeapp. The app is compatible with versions of Cytoscape 3.7 and above.
Source code available at: https://github.com/cannin/causalpath_cytoscape_app
Archived source code at the time of publication: https://doi.org/10.5281/zenodo.608165913
License: Apache License 2.0 license.
We thank Scooter Morris and Alexander Pico for their valuable feedback regarding Cytoscape during the development of the project. Additionally, we would like to thank the coordinators of the Google Summer of Code program.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Systems biology; Omics integration; Metabolic regulation
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics
Is the rationale for developing the new software tool clearly explained?
Yes
Is the description of the software tool technically sound?
Yes
Are sufficient details of the code, methods and analysis (if applicable) provided to allow replication of the software development and its use by others?
Partly
Is sufficient information provided to allow interpretation of the expected output datasets and any results generated using the tool?
Partly
Are the conclusions about the tool and its performance adequately supported by the findings presented in the article?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 2 (revision) 14 Nov 22 |
read | read |
|
Version 1 25 Apr 22 |
read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)