Keywords
Cannabis Sativa, THCAS, molecular cloning, bioinformatics, modeling.
This article is included in the Plant Science gateway.
This article is included in the Bioinformatics gateway.
Cannabis Sativa, THCAS, molecular cloning, bioinformatics, modeling.
In this study, we cloned the THCAS gene from Moroccan cannabis into an expression vector PpinkαHC, to transform the E.coli bacteria, for future expression in the yeast Pichia pasteri. We explored the protocol of molecular cloning of the recombinant THCAS gene for the Moroccan variety Cannabis sativa. and then we predicted three-dimensional structure of the recombinant enzyme THCAS exploiting deeplearning technology and the Rosetta Comparative Modeling (RosettaCM) method of the ROBETTA server.
In the first version, all the molecular cloning steps were submitted in detail except for the step of ligation of the enzymatic digestion products (the plasmid and the gene of interest), as well as the bacterial transformation, these steps were briefly mentioned, in order to meet the requirements of the reviewers. Moreover, the reviewers pointed out modifications that I have taken into consideration. The second version includes these two steps in detail, with the modifications related to the reviewers' revision, including a new Figure 6.
See the authors' detailed response to the review by Noureddine Hamamouch
See the authors' detailed response to the review by Supaart Sirikantaramas
Cannabis sativa L., basically originated from Central Asia, is the most widely used illegal drug around the world, appallingly consumed by around 160 millions individual. It is certainly the most popularly cultivated with two main derivatives, which are marijuana and resin.1 In fact, Morocco remains the world’s leading producer of cannabis resin as declared by the United Nations Office on Drugs and Crime (UNODC).2 Remarkably, since the psychoactive cannabinoid tetrahydrocannabinol (THC) was first discovered in 1964 and the human endocannabinoid system in 1980, many studies focused on the therapeutic effects and pharmaceutical applications of THC. Currently, the active ingredient THC is increasingly used in the treatment of nausea and vomiting associated with chemotherapy, loss of appetite due to AIDS, pain, and muscle spasms because of multiple sclerosis. As of today, Sativex, a cannabis-based preparation containing THC, was licensed in Canada as a neuropathic analgesic for multiple sclerosis.3 Furthermore, many other applications are still being explored, and demand for Pharma-Grade THC continues to grow.4 The heterologous production of cannabinoids or their precursors has been attempted in different microorganisms, such as E. coli,5–9 Zymomonas mobilis,10 Synechocystis spp.11 and Synechococcus elongatus.12 Besides, fungal species like Saccharomyces cerevisiae,13–17 Kluyveromyces marxianus,13 Komagataella phaffii,14,18,19 Candida viswanathii,20 or plant species like Chlamydomonas reinhardtii12 and Nicotiana benthamiana were also exploited.21–23 E. coli was not a suitable host for expression of the recombinant THCAS enzyme. Experiments to produce THCA in methylotrophic yeast showed promising results using a Pichia pastoris expression system. In addition, recombinant THCAS purified from P. pastoris had a much higher activity level than native THCAS purified from C. sativa and recombinant THCAS produced by insect cells. However, the expression rate of recombinant enzymes is still insufficient for industrial expression.
New disquisitive approaches have been applied for a different line of genetic exploration on cannabis that described the sequence of Δ9-tetrahydrocannabinolic acid-synthase (THCAs), which is the enzyme behind the production THC from the cannabigerolic acid (CBGA) precursor.24 Indeed, the chemical production of THC has many limitations since the legal regulation of C. sativa cultivation is restricted in the majority of countries, the low yield of the asymmetric chemical synthesis of THC, and the difficult extraction of THCA. Fortunately, CBGA is easy to synthesize25 and THCA is naturally decarboxylated by simple heating or during storage.24 Therefore, a biotechnological approach could be a suitable alternative. Notably, the public server Robetta has been very efficient in protein structure prediction from the amino acid sequence. It has continuously demonstrated its reliable performance and accuracy in the CASP (Critical Assessment of Structure Prediction tests). It has been frequently evaluated by CAMEO (Continuous Automated Model EvaluatOn) since 2014.26 Moreover, the CASP10 experiment suggests that Rosetta Comparative Modeling (RosettaCM) produces models with more accurate side chain and backbone conformations compared to other methods especially when the sequence identity with the models is greater than 15% and the reference crystal structure is available. Furthermore, The complete coding region of the THCAS gene has been sequenced from DNA extracted from the resin of Moroccan cannabis.27 As of today, THCAS was only expressed in low amounts either in hairy roots, insect cells (Spodopterafrugiperda), or yeasts like Pichia pastoris (Komagataella), and Saccharomyces cerevisiae.25,28,29 Luckily in 2015, bioconversion of Pichia pastoris yeast cells led to the production of 1 mM of THCA (0.36g THCA l-1).14,30 However, cloning of THCAS gene hasn’t been yet performed in Morocco.
In order to produce the active ingredient THC in vitro, we aim to clone the THCA synthase gene extracted from Moroccan cannabis to get the recombinant enzyme, using the expression system Pichia Pastoris. We have covered in this present study the first part of our work; we isolated the THCA synthase gene from the leaves of the Moroccan variety C. sativa (khardala) without the native signal sequence, and then predicted the three-dimensional structure of the recombinant THCAS enzyme.
Raw, fresh C. sativa plant of the variety “Khardala” were collected from Fez in northern Morocco (N 34.036466, W 5.017336). By members of the gendarmerie, and sent for scientific investigation to the Genetics Analysis Institute of the Royal Gendarmerie, Rabat, Morocco.
First, Cannabis leaves (50 mg per tube) were ground by the Tissulyser II. Then, DNA was extracted by the ISOLATE II DNA Plant kit following the manufacturer’s instructions. Next, DNA was quantified using the NanoDrop 8000 spectrophotometer (ThermoFisher)31; one unit of absorbance (260 nm) was assumed to correspond to 50 ng of DNA per μl of solution.
We aligned the THCA synthase sequence isolated from Moroccan Cannabis resin, which has the GenBank accession number ((JQ437481)27 to the cDNA sequence obtained from the open reading frame (ORF) mRNA precursor of the THCAS gene, with the GenBank accession number (AB057805),32 deposited in NCBI.33
We deduced the specific primers of THCAS gene of the Cannabis Moroccan variety without the native signal sequence respecting the following conditions: (1) Specificity; (2) Compatibility; (3) Primer stability; (4) Primer length should be 15-30 bp; (5) Primer melting Temperature (Tm) of about 48 to 60°C; (6) Forward and Reverse primers’ Tm preferably in the range of 52-58°C; and (7) an optimum GC content ranging from 40 to 60%. These characteristics were tested by AmplifX34 and SnapGene35 softwares. The pairs of primers obtained were evaluated for non-specific hybridization to other regions of the genome using the Basic Local Alignment Search Tool (BLAST). The Multiplex Manager v.1.236 software was used to verify the primer-primer interaction, avoiding potential primer-dimers and secondary hairpin structures. The heterolog expression of Pichia pastors may be intracellular or secreted. Secretion requires the presence of a signal sequence on the expressed protein to target it to the secretory pathway. While, The secretion signal sequence from the Saccharomyces cerevisiae α-mating factor pre-sequence has been used with the most success than the native secretion signal present in proteins of interest. It means, that the secreted heterologous protein serves as the first step in the purification of the protein recombinant.
We used the same primers with adapter sequences for sub-cloning (Table 1). We first added the specific restriction site to the StuI enzyme to the sense primer. Then, three histidine codons, two transcription stop codons, and the restriction site specific to the KpnI enzyme to the antisense primer. The gene already contains 3 codon his in position the sens primer, we wanted to add the three histidine codon to simplify the purification step with chromatography after expression of the gene to the yeast. We amplified the large chain ribulose-bisphosphate carboxylase (rbcL) gene (rbcL sense primer: 5′-ATGTCACCACAAACAGAGACTAAAGC-3′; rbcL antisense primer: 5′-GCAGCAGCTAGTTCCGGGCTCCA-3′) for the PCR positive control.37 The last step was to predict the three-dimensional structure of the recombinant enzyme THCAS exploiting deeplearning technology and the Rosetta Comparative Modeling (RosettaCM) method of the ROBETTA server.
PCR amplification was performed following the standard protocol: preheating at 94°C for 5 minutes, followed by 35 cycles at 94°C for 1 minute, then at 55°C for 1 minute, and at 72°C for 2 minutes, with a final extension at 72°C for 10 minutes. The reactions were carried out in a BIO RAD S1000 thermal cycler.27
The amplified fragments were subjected to 1% of the agarose gel detected using ethidium bromide. Gels were visualized under UV trans-illuminator using the G:BOX gel system (Syngene).
We used two restriction enzymes for the digestion reaction; StuI (Sigma Aldrich), and KpnI (Roche Applied Science), which was successively performed in the L buffer from (Roche Applied Science). The first restriction enzyme reaction (StuI enzyme) was achieved using a final volume of 25 μl under the following conditions: 2.5 μl of 10X SuRe/Cut Buffer L, BSA final concentration of 100 μg/ml, and 1 μl of enzyme StuI 10 U/ul and 500 ng of DNA. The reaction was incubated for 1 hour at 37°C and inactivated by incubation for 20 min at 65°C. This was followed by a second restriction adding 1 ul of the enzyme KpnI 10 U/ul. The reaction was incubated for 1 h at 37°C and inactivated by incubation for 15 min at 65°C.
Genetic individualization of THCAS gene without it’s signal sequence facilitated integration of the insert into the plasmid of choice; pPinkαHC. Thus, the amplified amplicon by two primers including adaptors was digested in addition to the vector using compatible restriction enzymes. The digested insert and vector were joined by the ligase enzyme. Actually, this reaction is primordial to succeed the sub-cloning using the chemo-competent E. coli Dhα bacterium, and hence, express the THC gene of interest in a host like the yeast Pichia pastoris.
We illustrated in Figure 1 the cDNA of the THCAS gene performed by SnapGene software in order to specify the main parts that made up the sequence of the gene in question. In addition, Figure 2 shows the alignment of THCAS gene, with the genbank accession number (JQ437481) against its formerly sequenced cDNA precursor’s counterpart, with the Genbank accession number (AB057805). The alignment result showed a high similarity of 98% starting from ATG star codon to the last three CAT (Histidine) codons of the THCAS gene.
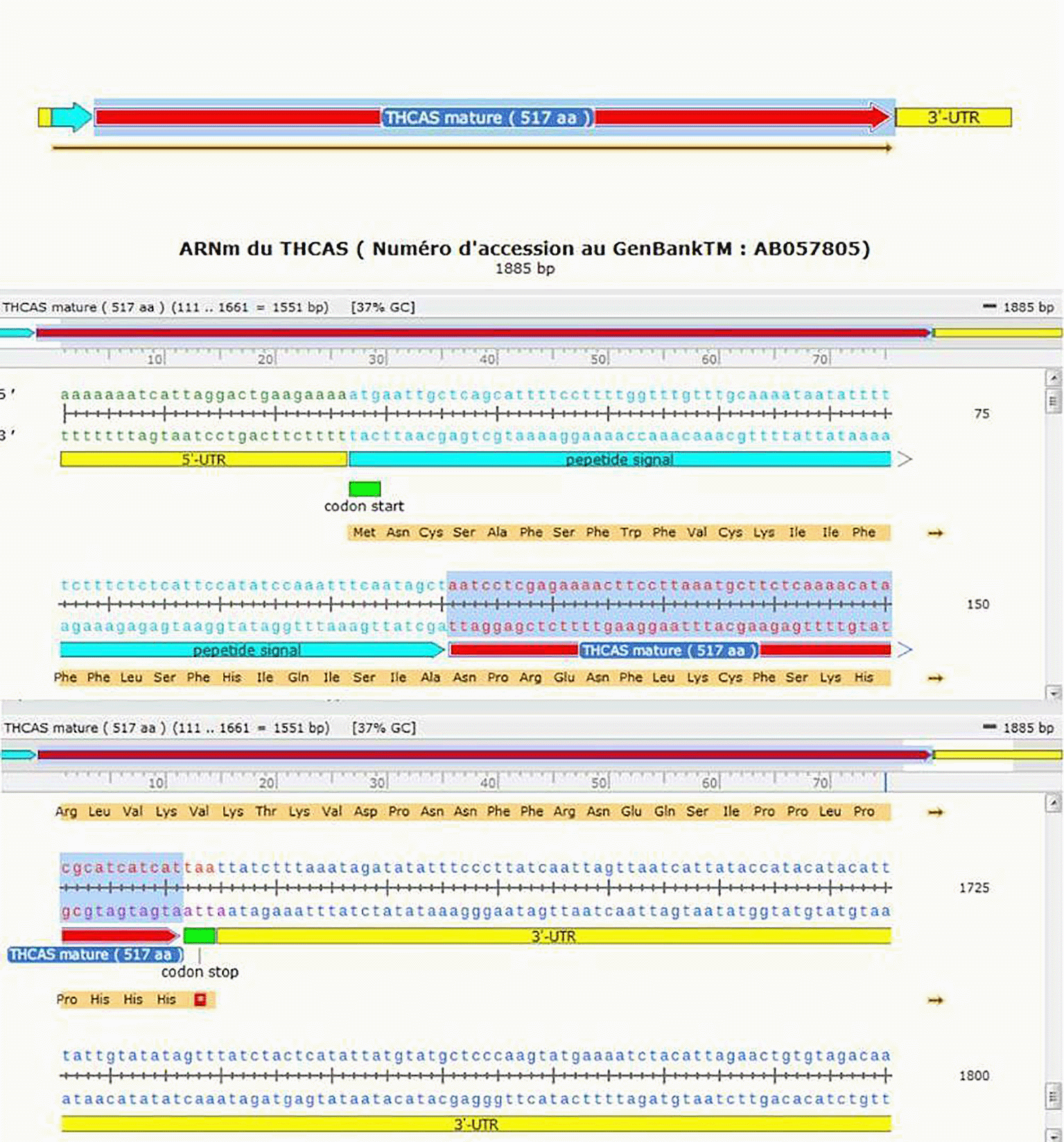
Legend: 5′UTR: Untranslated Transcribed Region 5′; 3′UTR: Untranslated Transcribed Region 3′.
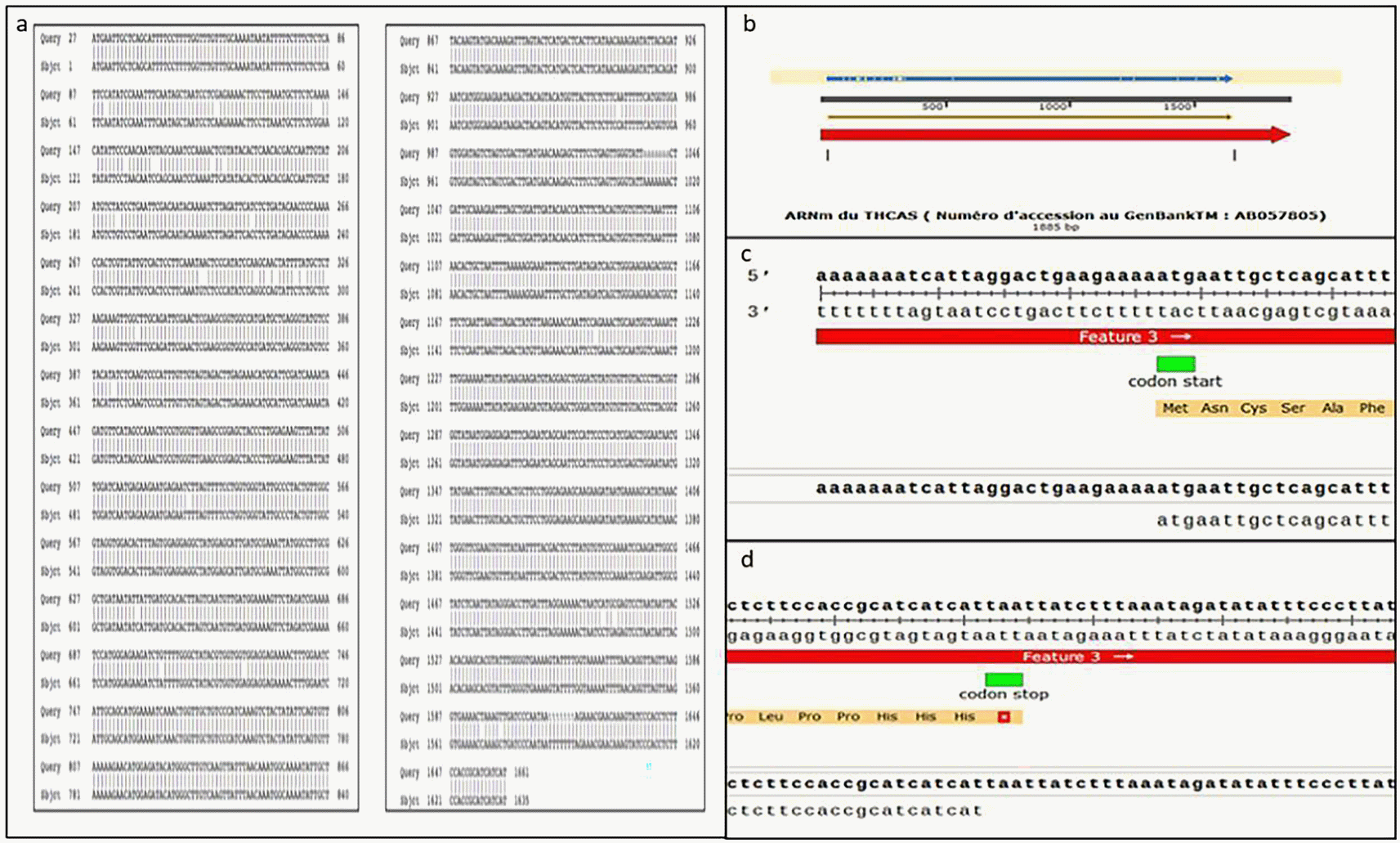
Legend: Query: C. sativa mRNA for tetrahydrocannabinolic Acid synthase precursor (AB057805), which is 1885 nucleotides long; Subject: Cannabis sativa isolate 01 tetrahydrocannabinolic acid synthase gene (JQ437481), which is 1635 nucléotides long; b) Alignement of the THCAS gene extracted from Moroccan Cannabis sativa with accession (JQ437481) marked in blue in relation to cDNA of mRNA with accession (AB057805) marked in Red using SnapGene software; c) Start codon (5′ ATG); Stop codon (3′ TAA).
Figure 3 showed the different samples of harvested Moroccan C. sativa namely “Khardala” including duplicates to assess variance in the evaluation method, including both sampling and analysis. Samples are numbered from 1 to 6 including their weight measurement 100, 150, 200, 100, 50 and 50 mg, respectively. In addition, this figure showed the summary of DNA quantification, which varied between 75 ng/ul and 1279 ng/ul, with a ratio of 260/280 and 260/230 between 1 and 2.5.
All PCR primers with and without adapters amplified a sequence of 1551 nucleotides, from position 85N to 1635N of the THCAS gene. Additionally, amplification of the rbcl cannabis-specific positive control was successful, while no amplification was observed at the negative control (water). This result was repeated several times for confirmation with regards to samples and duplicates (Figure 4).
Legend: g: amplification of DNA extract (THCAS gene); w: negative control amplification (water).
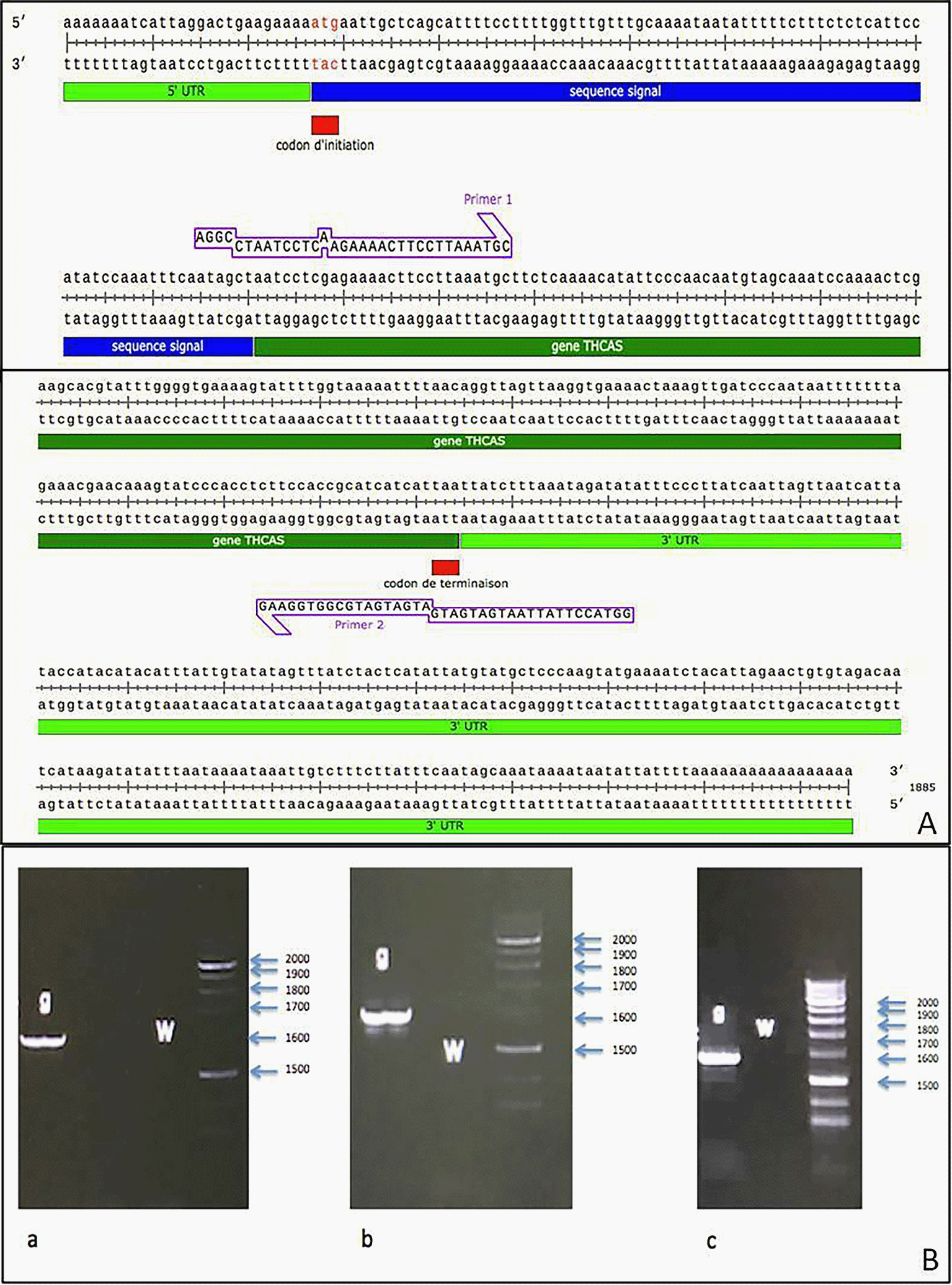
We added the forward and reverse primers to the Stul and Kpnl restriction enzymes, respectively. Histidine codons (ATG) and transcription stop codon (TTA) were added to the reverse primer. On the one hand, the enzymatic restriction was carried out in a successive manner and in both directions at the level of the pPinkα-HC vector. On the other hand, the same enzymatic restriction reactions were carried out for the amplicon produced with the primers with adapters (Figure 5).
A) Vector named pPinkα-HC is composed of: 7898 nucleotides; α-factor: the signal sequence; MSC: multiple cloning site (Stu I, Kpn I, Nae I, Fse I or Swa I); 5′ AOX1 promoter region (alcohol oxidase promoter); CYC1 TT: Cytocrome C1 transcription termination; PUC ori: PUC origin of replication; AmpR: ampicillin resistance gene; P ADE2 HC: The ADE2 promoter region; ADE2: Open reading frame ADE2 (responsible for the synthesis of Adenine) TRP2: Gene TRP2 (responsible for the synthesis of tryptophan).
B) Visualization on agarose gel of the pPinkα-HC vector after enzymatic digestion; 1) 1 Kb ladder; 2) undigested plasmid; 3) digested plasmid with the StuI enzyme; 4) digested plasmid with StuI enzyme, followed by the KpnI enzyme; 5) Digested plasmid with KpnI enzyme; 6) digested plasmid with Kpnl enzyme, followed by the Stul enzyme;
C) Ppink-HC vector as illustrated by SnapGene software after enzymatic digestion.
D) Result of PCR amplification: The amplified THCAS gene without signal sequence, the StuI restriction site is present at the 5′ position, the KpnI restriction site at the 3′ position, using the snapgen software;
E) Revelation of THCAS gene without its signal sequence on agarose gel after the enzymatic digestion;
F) THCAS gene without its signal sequence after enzymatic digestion as shown by SnapGene software.
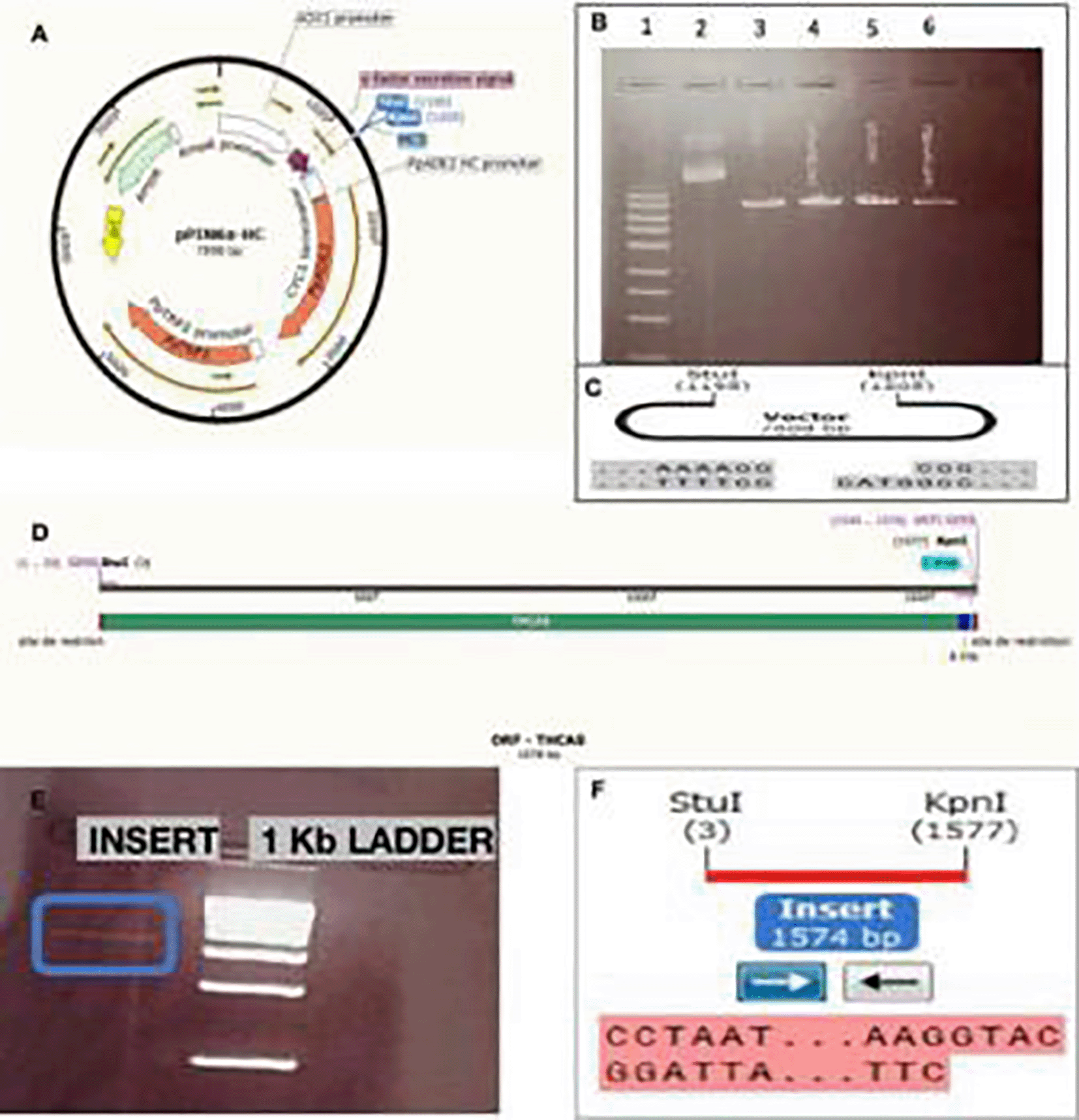
We performed the ligation of the vector and the THCAS gene without the signal sequence and after the enzymatic digestion step, followed by the transformation of the chemo competent E. coli DHα and the inoculation of solid agarose culture medium (Figure 6).
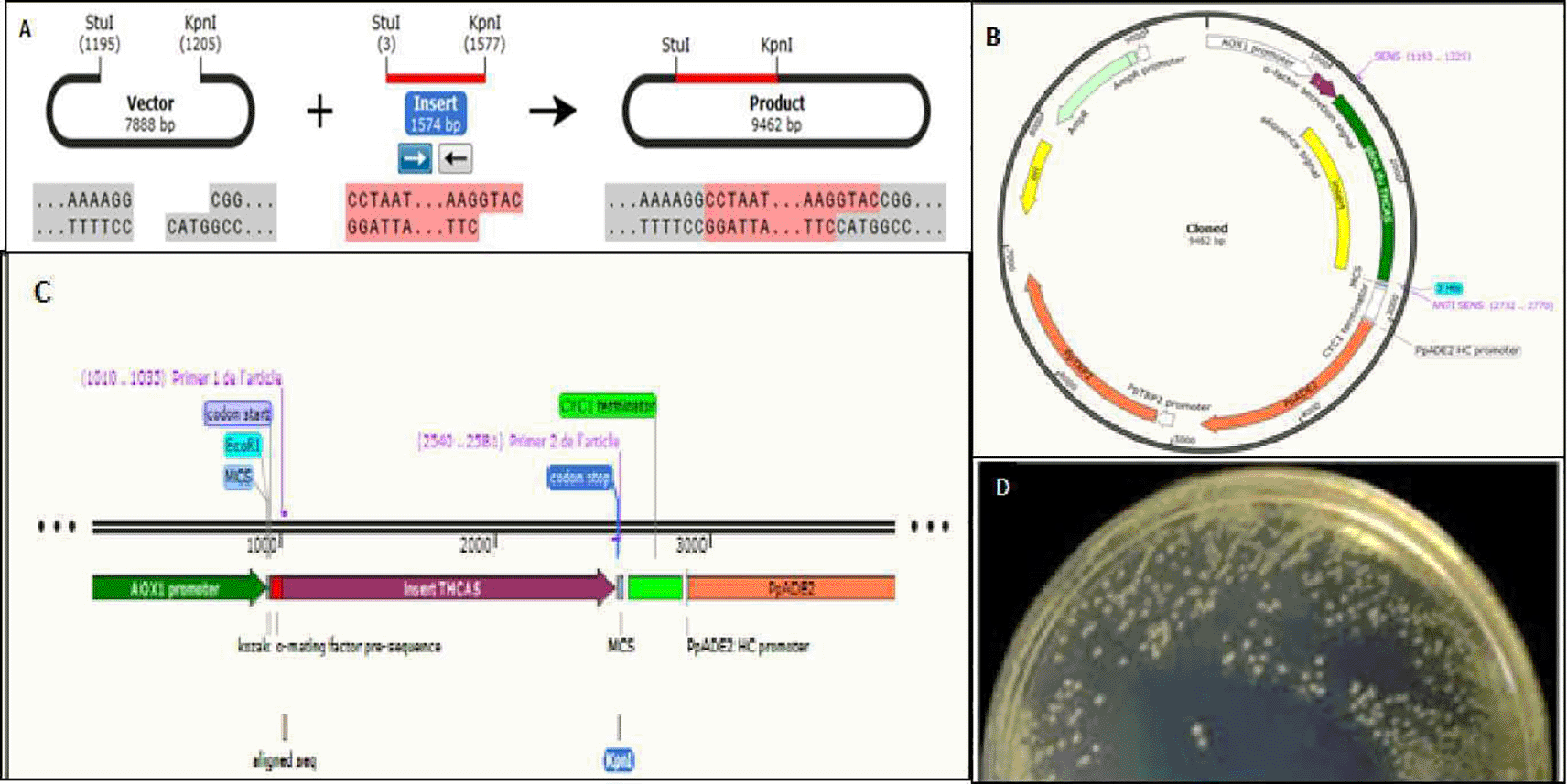
A) Ligation reaction. B) Result of insertion of THCAS gene without sequence signals to the vector pPinkα-HC complete. C) The part of THCAS gene without the sequence signals between the two sequences of the promoter and the terminator respectively illustrated by the SnapGene software. D) Result of culture of E. Coli DHα transformed bacteria.
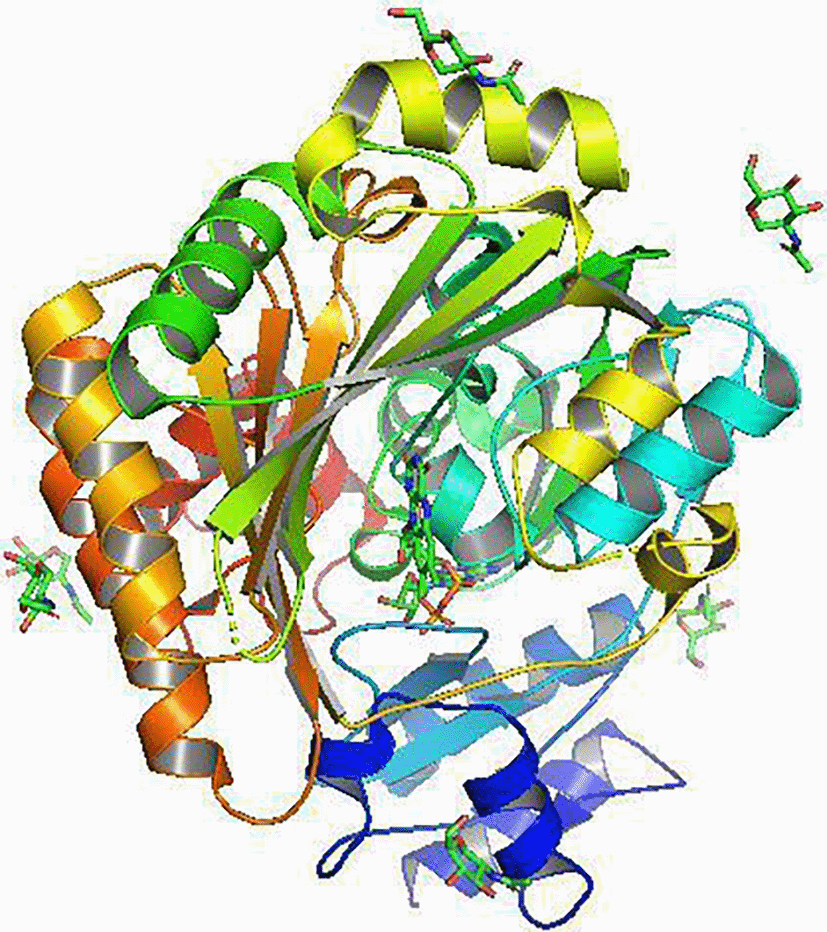
THCAS gene of Moroccan C. sativa was sequenced with accession (JQ437481-JQ437488), knowing that THCAS gene has a size of 1635 nucleotides,32 and Its signal peptide sequence is 28 amino acids. Following the alignment of THCAS gene of Moroccan C. sativa against its cDNA precursor, we observed that the former allows an open reading frame of 1635 nucleotides, coding for a polypeptide of 545 amino acids (Figure 7). Thus, the length of the predicted mature THCAS polypeptide is 517 amino acids (Figures 8 and 9). We identified the NH2-terminal sequence as: NPRENFLKXFSKHIPNNVANPKLV, which corresponds to the nucleotide sequence 84-156 Ns of THCAS gene (GenBank accession number: JQ437481). It has 9 discrepancies with the NH2-terminal sequence of the cDNA (GenBank accession number: AB057805). RMSDs (root mean square deviation of atomic positions) for C-alpha atoms, main chain atoms, side chain atoms and all atoms between the modelled structure and the model corresponding to 1.325 Å (Figure 9).

Usage of C. sativa for pharmacological purposes, regardless of persistent controversy, helped reemerging this plant with historical significance in medical field. Morocco has been and still one the world’s largest producer of cannabis resin, with an estimated open air production of 38000 tons, and in door production of 760 tons, which helped gaining a net of an estimated 9 Billion dollars.41 Legislation with regards to governing use of cannabis for medical purposes in Morocco continues to evolve promptly,42 which requires implication of scientists, doctors, and pharmacists in studying the healthy benefits of Cannabis. Moreover, THC has generated recently considerable interest for its new and valuable pharmacological activities, and hence, is been commercially available, but extremely expensive, which makes it difficult to be a target for detailed studies. Unfortunately, application of conventional methods to prepare THC, including organic synthesis and isolation from marijuana, were not very practical ways to provide enough pure THC.
In this study, we aimed to describe the recombinant THCAS gene cloning steps in order to exploit its recreational effect on the endocannabinoid system. Eventually, we sought to produce a recombinant enzyme with a specific and original enzymatic activity as well as a high production yield. To do so, we blasted the tetrahydrocannabinolic acid synthase (THCAS) gene isolated from the resin of Moroccan C. Sativa (JQ437481) corresponding to 1635 nucleotides against the mRNA precursor of THCAS (AB057805) Corresponds to 1885 nucleotides. Moreover, we prepared the nucleotide sequence of interest for subcloning the vector pPinkα-HC into E-Coli in order to express it in the yeast Komagataella phaffi (previously called Pichia pastoris). Ultimately, we predicted the final structure of the recombinant protein THCAS responsible for the production of the main active principle in Moroccan cannabis.
This work is the first step towards the expression of the recombinant THCAS enzyme in Komagataella phaffi yeast. We therefore hypothesized that recombinant THCA synthase would contribute to the biotechnological production of THC, once a suitable expression system is developed. since THCA is quantitatively decarboxylated to THC by heating. As a possible system, we have here developed the possibility of expressing recombinant THCAS in transgenic P. pastoris secreting THCA synthase. In order to achieve this goal, we started with the alignment of the THCAS sequence.
Sequence alignment helped identifying the conserved region that correspond to the position of key residues (his 114, cys176, his292, glu417, tyr484, Cys37 and Cys99), which are conserved in the THCAS of Moroccan C. sativa. These sites are essential for the enzymatic activity of THCAS, and found to be present at the same positions as previously described amino acids. THCAS protein, is a flavoprotein, since the presence of coenzyme FAD is essential for its enzyme activity.30 Besides, it covalently binds to His114 and Cys176.32 In addition, Tyr 484 being the catalytic site of the enzyme, including both His292 and Tyr417, are involved in binding to the CBGA substrate. the three-dimensional structure of the enzyme in question is generated thanks to the disulfide bond (between Cys37 and Cys99) and the six N-glycosylation sites.40 Interestingly, the alignment showed a 98% match, which implies a high quality. Therefore, it was not necessary to improve the alignment by correcting errors or eliminating gaps.
The yield of the extracted genomic DNA gave satisfactory, significant concentration, proportional to the mass of the initial sample. the purity of the extracted DNA was evaluated by the A260/230 ratio to assume the contamination level. The presence of polysaccharides and phenolic compounds, detergents and other salts might explain the A260/230 value, which is close to the ideal value of 2.31 The PCR quality suggests that the extract was pure enough for downstream applications, such as enzymatic DNA digestion and ligation. Meanwhile, a single band of approximately 1600 nucleotides was obtained after enzymatic restriction. Hence, we deduced that the choice of restriction enzymes was successful because they cut at the level of the THCAS gene of interest and recognized a restriction site composed of 6 nucleotides to maintain the same reading frame of the gene of interest. Furthermore, the three histidine codons were added to facilitate the extraction of the recombinant enzyme by HPLC chromatography.
In the first run, we performed enzymatic restriction with StuI and KpnI at the same time, because the two enzymes have 100% activity in the SuRe/Cut Buffer L. However, both enzymes lost their enzymatic activity, which implies that the presence of the two reacting enzymes initiates their activities. Consequentially, we carried out a restriction in a successive way and in both directions to ensure that the activity of each enzyme remains. Later, the same reactions were performed for the amplicon produced by the adapter primers. This result may be useful for applications like ligase reaction and molecular cloning, which means that primers design was successful. However, a sequencing step would be crucial for confirmation.
THCA synthase is identified as a monomeric enzyme. It consists of two domains (Domains I and II) divided by the FAD.40 The formation of the oxidative cyclization reaction will likely occur through intermediaries.43 Along the entire length of the THCAS enzyme protein, there are positions that interact with the CBGA substrate or the FAD cofactor. This leads us to the need for molecular cloning of the entire nucleotide sequence, as has been done previously.14,29,32 In addition to obtaining greater enzymatic activity of the recombinant enzyme. Endoglycosidase treatment yielded deglycosylated THCA synthase with higher catalytic activity than either the glycosylated form of the native enzyme or the recombinant enzyme.29
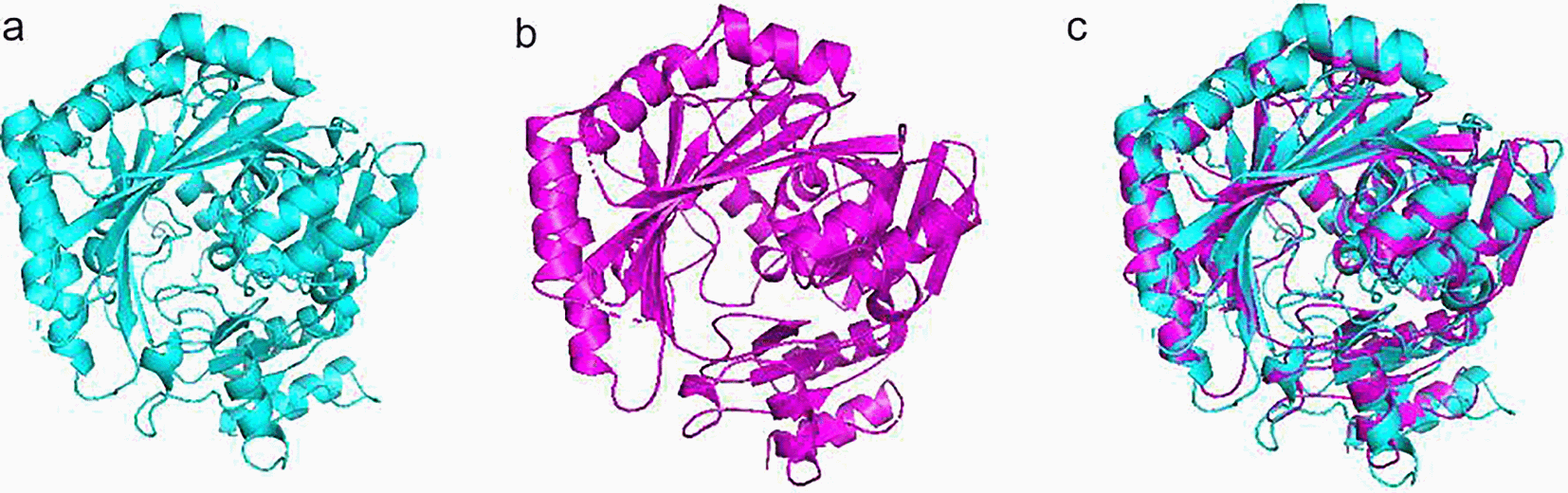
Furthermore, the RMSD of the modeled structures and the corresponding reference model is 1.325 Å, reflecting the high quality of the obtained models. This result is essential for earlier applications such as molecular cloning. DeepMind’s Alphafold2 demonstrated its remarkable robustness and accuracy for the example of our enzyme of interest with this result. Each possible change in the nucleotide sequence made by the site-directed mutation during molecular cloning can be tested in silico before carrying it out at the laboratory level by modeling in X-ray crystallography and cryo-electron microscopy and generating precise models of complexes. protein-protein and saved considerable time.26
Additionally, The Rosetta Comparative Modeling (RosettaCM) experiment produced models with more accurate side chain and backbone conformations than other methods when the sequence identity with the models is greater than 15% and the crystal structure of the model is arranged. Other works have been executed in this sense by the same THCAS gene of Moroccan origin using conventional techniques such as Modeling.44 Remarkably, the same modeling accuracy was approximately obtained.
Unfortunately, little is still known about THCAS in Cannabis, which constitutes a valuable unexploited treasure. However, artificial intelligence is being applied by pharmaceutical companies to discover drugs and design new optimized compounds in order to accelerate the identification of promising drug candidates for specific indications. Hence, we applied this technique on the THCAS of Cannabis sativa for the first time in Morocco. The use of the 3VTE reference structure helped improving both the number of aligned residues and the quality of the superposition, evaluated by the root mean square deviation of atomic position (RMSD) calculation. Furthermore, the modeled protein was superimposed to the reference structure to assess the predicted three-dimensional structure quality, which showed a good conservation of the model’s global folding. This would foster a better understand of the crucial conformation at the active principle level, which could explain the variation in the rate of THCA in the simulated model by bioinformatics. We ultimately aim to generate a molecular cloning protocol of THCAS gene of interest and hence, predict the most relevant recombinant enzyme for gene expression in yeast. Therefore, we will be saving time, energy and resources in laboratory work. However, this method remains incomplete to judge the reliability of the recombinant enzyme. It is advantageous to go through in vitro and in vivo experiments before any human drug testing.
This study demonstrated a great similarity between the THCAS gene of Moroccan Cannabis variety and the formerly reported cDNA precursor gene reference. We isolated the nucleotide sequence of the THCAS gene using targeting specifics primers in order to express this gene in an appropriate host. Thus, we were able to predict for the first time, the structure of the recombinant THCAS enzyme using in silico, in vitro, and artificial intelligence tools. These results are essential to successfully clone in a eukaryotic host and express the THCA recombinant enzyme in the yeast Pichia pastoris.
The THCAS sequences with a complete open reading frame sequenced from Moroccan cannabis resin are available under these Genbank accession numbers27:
GenBank: Cannabis sativa isolate 01 tetrahydrocannabinolic acid synthase gene, partial cds. Accession number: JQ437481.
GenBank: Cannabis sativa isolate 02 tetrahydrocannabinolic acid synthase gene, partial cds. Accession number: JQ437482.
GenBank: Cannabis sativa isolate 03 tetrahydrocannabinolic acid synthase gene, partial cds. Accession number: JQ437483.
GenBank: Cannabis sativa isolate 04 tetrahydrocannabinolic acid synthase gene, partial cds. Accession number: JQ437484.
GenBank: Cannabis sativa isolate 05 tetrahydrocannabinolic acid synthase gene, partial cds. Accession number: JQ437485.
GenBank: Cannabis sativa isolate 06 tetrahydrocannabinolic acid synthase gene, partial cds. Accession number: JQ437486.
GenBank: Cannabis sativa isolate 07 tetrahydrocannabinolic acid synthase gene, partial cds. Accession number: JQ437487.
GenBank: Cannabis sativa isolate 08 tetrahydrocannabinolic acid synthase gene, partial cds. Accession number: JQ437488.
cDNA of theTHCAS precursor mRNA in C. sativa sequenced in Japan, used for comparative bioninformatics analysis is available under the GenBank accession number32:
GenBank: Cannabis sativa mRNA for tetrahydrocannabinolic acid synthase precursor, complete cds. Accession number: AB057805.
I would like to express my sincere thanks to all co-authors. I am highly indebted to my supervisors for their guidance and constant mentoring as well as for providing necessary information regarding the study and also for their support in completing the project.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Partly
References
1. Taura F, Dono E, Sirikantaramas S, Yoshimura K, et al.: Production of Delta(1)-tetrahydrocannabinolic acid by the biosynthetic enzyme secreted from transgenic Pichia pastoris.Biochem Biophys Res Commun. 2007; 361 (3): 675-80 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Biochemistry and Molecular Biology of Plants
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Enzymology, Plant specialized structures. Essential oils, docking, In vitro characterization of enzymes
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
No
Competing Interests: Dr Noureddine Hamamouch is the PhD supervisor for Bouchra Chaouni
Reviewer Expertise: Plant Molecular Biology and Biotechnology.
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
No
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
No
References
1. Taura F, Dono E, Sirikantaramas S, Yoshimura K, et al.: Production of Delta(1)-tetrahydrocannabinolic acid by the biosynthetic enzyme secreted from transgenic Pichia pastoris.Biochem Biophys Res Commun. 2007; 361 (3): 675-80 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Plant biochemistry and molecular biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
|
Version 2 (revision) 15 Dec 22 |
read | read | ||
|
Version 1 27 Jul 22 |
read | read | ||
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)