Keywords
Malpighiales, Phyllanthaceae, phylogenetic relationships, plastome, star gooseberry
This article is included in the Plant Science gateway.
This article is included in the Genomics and Genetics gateway.
Phyllanthus acidus (L.) Skeels (Phyllanthaceae) is a potential medicinal plant recognized for its sour and tart-tasting fruits. In this study, the chloroplast genome of P. acidus was sequenced, assembled, and characterized. The chloroplast genome size was 156,331 bp, and the overall GC content was 36.9%. Additionally, the chloroplast genome had a quadripartite structure consisting of a large single copy (LSC; 85,807 bp in length; GC content: 34.6%), a small single copy (SSC; 19,262 bp in length; GC content: 30.6%), and two inverted repeat regions (IR; 25,631 bp in length; GC content: 43.1%). A total of 113 unique genes were annotated in the chloroplast genome, including 79 protein-coding genes, 30 tRNAs, and four rRNAs. The phylogenetic analysis based on 79 protein-coding genes revealed the paraphyly of the Phyllanthus genus. These findings provided additional genetic information for further research on P. acidus and the cp genome in the Phyllanthaceae family.
Malpighiales, Phyllanthaceae, phylogenetic relationships, plastome, star gooseberry
The revised manuscript was modified based on the comments of two new reviewers about the content of version 2 related to the aim of the study, the tool (OrgabellgenomeDRAW which is for illustrating the genome map) for annotating chloroplast genome, and the method to extract protein-coding genes for phylogenetic analysis. We revised the aim of the study and added more details about the methods of our manuscript (e.g., annotating, illustrating, and extracting sequences from chloroplast genomes). For the comment about phylogenetic analysis, we added an additonal description about the phylogenetic result and refused to add discussion because of the regulation of Genome Note regulation. There is no change in title, figure, table, and authors of the manuscript.
See the authors' detailed response to the review by Jessica D. Rey
See the authors' detailed response to the review by Qiang Li
See the authors' detailed response to the review by Shiou Yih Lee
Phyllanthus acidus (L.) Skeels, also known as star gooseberry, belongs to the Phyllanthaceae family and is commonly distributed in the wet tropical regions, including South Asia, Southeast Asia, Central Africa, the Caribbean region, Central America, and South America (POWO 2022). P. acidus has been traditionally used to treat various diseases, including inflammation, gastrointestinal problems, rheumatism, bronchitis, Alzheimer’s, and hepatic diseases (Jain et al. 2011; Chakraborty et al. 2012; Srirama et al. 2012; Uddin et al. 2016). The leaves and roots of P. acidus also possess antidotal properties against viper venom (Jayvir 1998). Moreover, P. acidus could potentially alleviate hypertension (Leeya et al. 2010).
The chloroplast (cp) genome is highly effective at inferring phylogeny since it is predominantly maternally inherited, has a conversed structure and gene content, and has slow mutation frequency (Palmer et al. 1988). Additionally, the cp genomes provide essential data for examining population genetics, molecular identification, and genetic engineering (Powell et al. 1995; Daniell et al. 2016; Cao et al. 2022). Previously, a comparative chloroplast genomic study revealed the features of Phyllanthus chloroplast genomes and the non-monophyletic status of Phyllanthus species (Fang et al. 2023). Therefore, in the current study, we explored the characteristics of the P. acidus cp genome and its phylogenetic implication to gain more genomic information about the chloroplast genome evolution and to provide more data to resolve controversy of phylogenetic relationships within the Phyllanthus genus and closely related taxa in Phyllanthaceae.
The P. acidus sample (young branches with leaves) was collected from Can Tho, Viet Nam (9°56′55.7″N, 105°30′16.0″E) and labeled with voucher number: NTT-2022.12.CR (contact person: Dr. Do Hoang Dang Khoa, dhdkhoa@ntt.edu.vn). It was deposited at the NTT Hi-tech Institute, Nguyen Tat Thanh University. No specific permit was required to collect and study the species in Vietnam. The leaf sample was dried with silica gel and stored in a -80°C freezer until DNA extraction was conducted.
The total genomic DNA extraction from the dried leaves was carried out using the Cetyltrimethylammonium bromide (CTAB) protocol (Doyle & Doyle 1987). The quality of genomic DNA samples was checked using gel electrophoresis and the NanoDrop OneC Spectrophotometer. The DNA samples that showed a clear band on agarose gel and had a 260/280 ratio between 1,8-2 and a 260/230 ratio between 2.0-2.2 were selected for conducting the next-generation sequencing step. Subsequently, the library was prepared with the NEBNext Ultra II DNA Library Prep Kit for Illumina (NEB, USA). The library was sequenced using the Illumina MiSeq platform to generate paired-end reads of 150 bp (Ktest Science Co. Ltd., Vietnam). The raw reads were qualified and filtered for low-quality reads (Q score < 20 and length < 100 bp) and reads containing primers or adapters using FastQC v0.12.1 and Trimmomatic v0.39 programs (Andrews 2010; Bolger et al. 2014). For the assembly of the cp genome, the NOVOPlasty v4.3.1 program was used (Dierckxsens et al. 2017). Preliminary annotation was conducted by Geseq with default parameters for land plant plastid and chloroplast land plant reference set (MPI-MP Reference Set) (Tillich et al. 2017). The complete annotation genome was illustrated using OrganellarGenomeDRAW v1.3.1 with default settings for circular plastid genome (Greiner et al. 2019). All 79 protein-coding regions in the cp genomes of P. acidus and 16 related taxa from the Phyllanthaceae were extracted and aligned for phylogenetic analysis using MUSCLE v5 program (Edgar 2004). The chloroplast genome of Acalypha hispida (Euphorbiaceae; Genbank accession no. NC_070339) was selected as an outgroup. A maximum likelihood phylogenetic tree was reconstructed using IQTREE with 1000 bootstrap replicates and GTRGAMMA substitution model (Nguyen et al. 2015). In addition, Bayesian inference method was applied to the aligned sequnces under GTRGAMMA substitution model using MrBayes v3.2.7a with 1,000,000 generations and a discard of 25% sample (Ronquist et al. 2012).
Approximately 349.8 MB of clean reads were obtained and used for completing the cp genome of P. acidus. The assembly process utilized 1,166,034 paired-end reads, resulting in an average coverage depth of 2,234.3X (Nguyen, Nguyen, Do, and Vu, 2023; Nguyen, Do, and Vu, 2023) (Supplementary Figure S1, Do 2025). The quadripartite cp genome of P. acidus (GenBank accession number OR050568) had a length of 156,331 bp and consisted of an LSC region of 85,807 bp, a SSC region of 19,262 bp, and a pair of IR regions of 25,631 bp ( Figure 1). The overall GC contents of the genome was 36.9%, and the GC content of the LSC, SSC, and IR regions were 34.6%, 30.6%, and 43.1%, respectively. The cp genome of P. acidus contained a total of 130 genes, including 85 protein-coding regions, 37 tRNA genes, and eight rRNA genes (Table 1). Among 85 protein-coding genes, 17 genes contained introns, of which ycf3 and clpP contained two introns (Supplementary Figure S2, Do 2025). In IR regions, a total of 19 genes were duplicated, including eight protein-coding regions (i.e., rps19, rpl2, rpl23, ycf1, ycf2, ndhB, rps12, and rps7), seven tRNAs (trnI_CAU, trnL_CAA, trnV_GAC, trnI_GAU, trnA_UGC, trnR_ACG, and trnN_GUU), and four rRNAs (rrn16S, rrn23S, rrn4.5S, and rrn5S). Notably, rps19 and ycf1 duplications were incomplete. The phylogenetic analysis revealed a paraphyly of Phyllanthus species, in which Breynia futicosa and Glochidion chodoense formed a clade with Phyllanthus amarus ( Figure 2). Therefore, more genomic data and samples of Phyllanthaceae species are required for further phylogenetic studies.
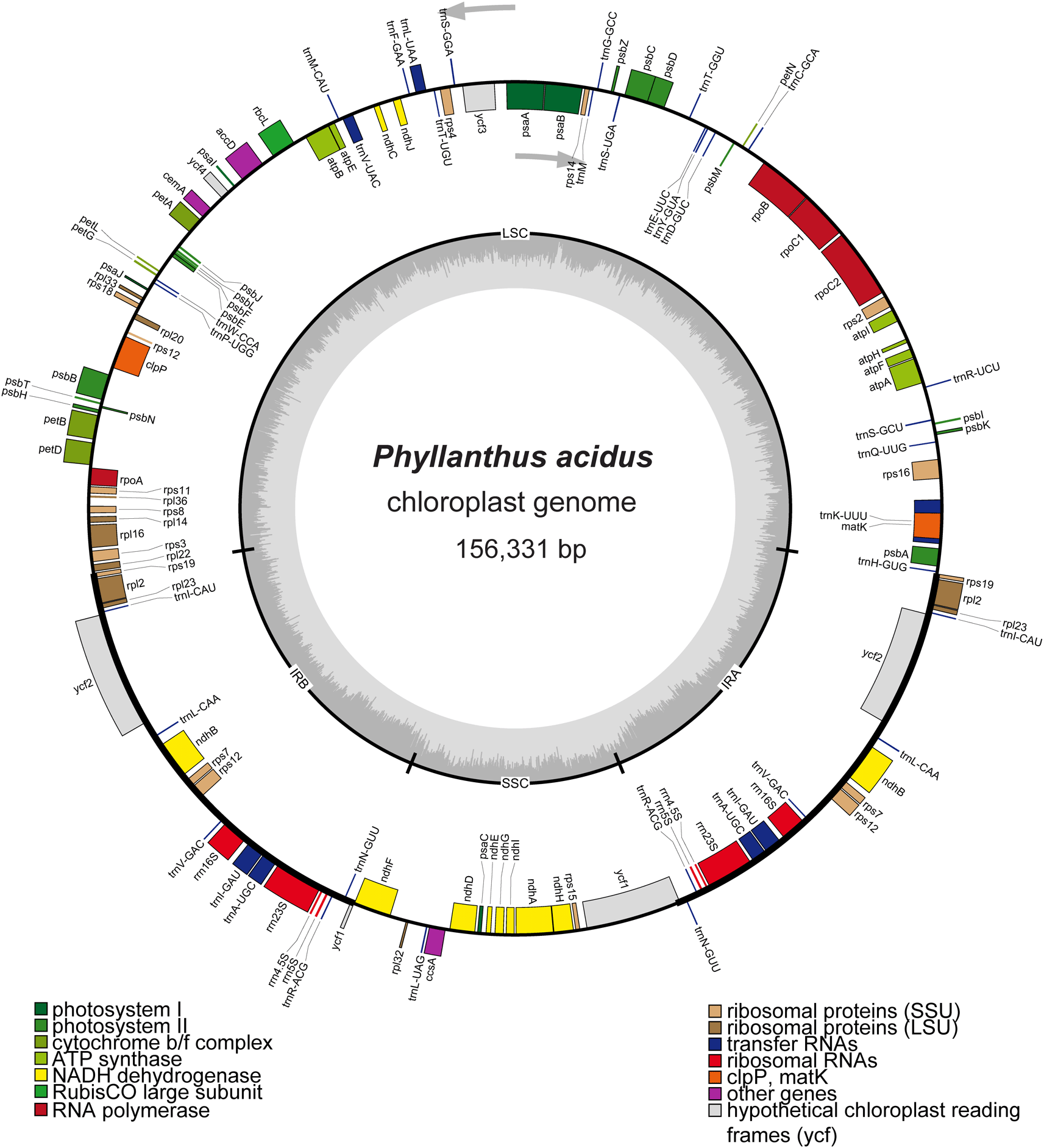
Genes located inside the circle are transcribed in a clockwise direction, while genes outside the circle are transcribed counterclockwise. The inner circle depicted in dark gray represents the GC content, while the light-gray circle represents the AT content of the genome. LSC: large single copy; SSC: small single copy; IRA/IRB: inverted repeat regions.
| Groups of genes | Name of genes |
|---|---|
| Ribosomal RNAs | rrn4.5(2x), rrn5(2x), rrn16(2x), rrn23(2x) |
| Transfer RNAs | trnA_UGC *(2x), trnC_GCA, trnD_GUC, trnE_UUC, trnF_GAA, trnG_UCC *, trnG_GCC, trnH_GUG, trnI_GAU *(2x), trnK_UUU *, trnL_CAA(2x), trnL_UAA *, trnL_UAG, trnfM_CAU, trnM_CAU(2x), trnM_CAU, trnN_GUU(2x), trnP_UGG, trnQ_UUG, trnR_ACG(2x), trnR_UCU, trnS_GCU, trnS_GGA, trnS_UGA, trnT_GGU, trnT_UGU, trnV_GAC(2x), trnV_UAC *, trnW_CCA, trnY_GUA |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| Cytochrome | petA, petB *, petD *, petG, petL, petN |
| ATP synthases | atpA, atpB, atpE, atpF *, atpH, atpI |
| Large unit of Rubisco | rbcL |
| NADH dehydrogenase | ndhA *, ndhB *(2x), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| ATP-dependent protease subunit P | clpP * |
| Envelop membrane protein | cemA |
| Large units of ribosome | rpl2 *(2x), rpl14, rpl16 *, rpl20, rpl22, rpl23(2x), rpl32, rpl33, rpl36 |
| Small units of ribosome | rps2, rps3, rps4, rps7(2x), rps8, rps11, rps12 *(2x), rps14, rps15, rps16 *, rps18, rps19(2xa) |
| RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 |
| Initiation factor | infA |
| Other genes | accD, ccsA, matK |
| Hypothetical proteins and conserved reading frames | ycf1(2xa), ycf2(2x), ycf3 *, ycf4 |
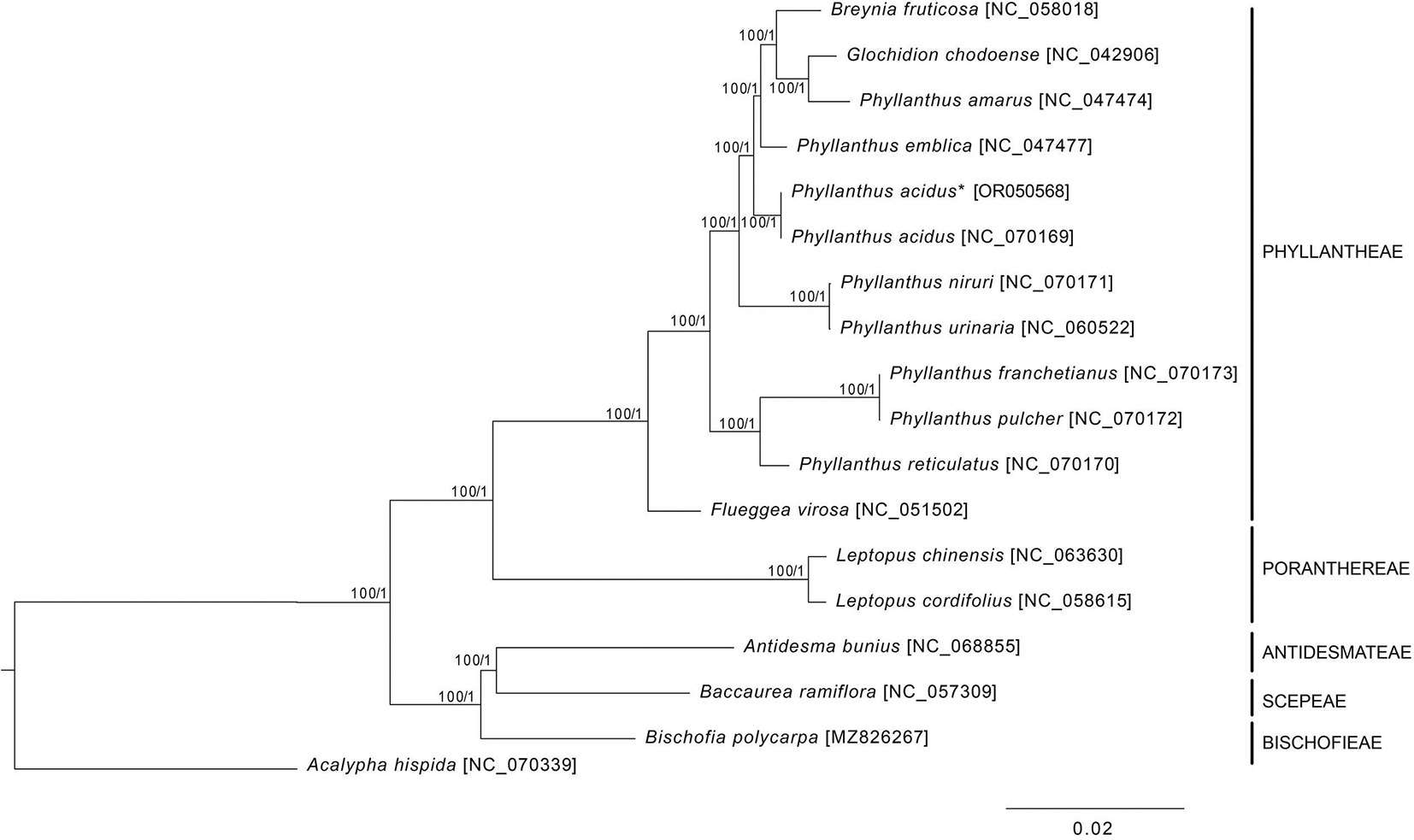
The asterisk indicates P. acidus sequenced in this study. The numbers next to each node are bootstrap values and posterior probability inferred from maximum likelihood and Bayesian inference methods.
NCBI Short Read Archive (SRA): DNA-seq of Phyllanthus acidus. Accession number SRR24772537; https://www.ncbi.nlm.nih.gov/sra/SRR24772537 (Nguyen, Nguyen, Do, and Vu, 2023).
NCBI Assembly database: Phyllanthus acidus chloroplast, complete genome. Accession number OR050568; https://www.ncbi.nlm.nih.gov/nuccore/OR050568 (Nguyen, Do, and Vu, 2023).
Figshare: Supplementary Files. https://doi.org/10.6084/m9.figshare.30354073 (Do 2025).
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0)
The authors thank to anonymous reviewers for their helpful comments to improve the quality of this manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Plant genomics and evolution
Are the rationale for sequencing the genome and the species significance clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Bioinformatics, genome assembly, and computational genomics.
Competing Interests: No competing interests were disclosed.
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: forestry, molecular phylogenetics and evolutionary
Are the rationale for sequencing the genome and the species significance clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
References
1. Nguyen H, Nguyen T, Vu M, Do H: The complete chloroplast genome of Phyllanthus acidus (L.) Skeels (Phyllanthaceae). F1000Research. 2025; 12. Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: My research areas primarily encompass speciation, phylogenetics, and the conservation of endangered plants. Currently, I mainly utilize chloroplast genomes, mitochondrial genomes, and nuclear genomes to address evolutionary questions.
Are the rationale for sequencing the genome and the species significance clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular characterization of different plant species i.e. Philippine endemic medicinal plants, strawberry varieties planted in the Philippines, genetics of disease resistance cultivated rice, soilless culture of leafy and fruiting vegetables
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |
|
Version 3 (revision) 21 Nov 25 |
read | read | read | ||
|
Version 2 (revision) 16 Oct 25 |
read | read | |||
|
Version 1 31 Aug 23 |
read | ||||
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)