Keywords
Breast cancer, IFN-γ, Hormonal, ER, PR, Triple negative, Immune cells
This article is included in the Oncology gateway.
This article is included in the Cell & Molecular Biology gateway.
Breast cancer, IFN-γ, Hormonal, ER, PR, Triple negative, Immune cells
Cancer is still the second leading cause of death in the world despite advances in cancer therapy and the emerging relevance of genomics in cancer treatment and precision medicine.1,2 Among all types of cancer, breast cancer (BC) is the most frequent type in women worldwide. According to the World Health Organization, each year, this malignancy affects more than two million women globally and is the primary cause of cancer-related death in women.3,4 Disease heterogeneity in BC increases its resistance to targeted treatment. BC can be molecularly classified into luminal A, luminal B, human epidermal growth factor receptor 2 (HER2) positive, and basal-like, with respect to the biomarkers status of estrogen receptor (ER), progesterone receptor (PR), and HER2.5,6 Particularly, the triple negative BC subtype is considered to be the most aggressive subtype and has poor outcomes due to limited options for effective treatment.7 However, there is a better overall survival (OS) rate for patients with ER+ or PR+ tumors, in part because of the intrinsic characteristics of these tumors and in part because of the improved treatments targeting these tumors.8 Approximately, 60% of diagnosed BC cases are ER+/PR+, while those expressing HER2 account for 15-20% of cases.9 In addition, the tumor microenvironment (TME) has a crucial role in tumor development and immune modulation. The TME may induce immune cells such as natural killer (NK) and T cells to release molecules that favor tumor progression and immune inhibition.10,11 However, the modulatory interactions between the tumor and the immune system remain poorly understood.
As is well known, immune cells get activated when encountering altered self cells including transformed and tumor cells. Activated immune cells secrete cytokines and chemokines, such as IL-12, IL-15 and interferon gamma (IFN-γ), to activate the immune cells besides its direct effect on the transformed cells.12,13 Recent data showed that IFN-γ is also involved in promoting the progression of different tumors with immunosuppression phenotype, which may be the reason for some patients showing resistance to immunotherapy.14 Furthermore, IFN-γ autocrine signaling has been detected in BC cells, affecting their behavior and sensitivity to treatment, besides suppressing tumor immune cells.15,16 Therefore, understanding the role of IFN-γ in defining tumor heterogeneity may advance our knowledge of TME and allow for more effective personalized immunotherapeutic approaches in BC.
This study was performed on the triple negative human BC cell lines (TNBC) MDA-MB-231(ER-, PR-, HER2-) (RRID:CVCL_0062) and transformed hormone receptor-positive MDA-MB-231 (ER+, PR+). Triple negative and ER- and PR-positive cells were then treated with IFN-γ, followed by sequencing and analysis using different bioinformatics tools. Figure 1 summarizes the experimental methods and software analyses used in this study.
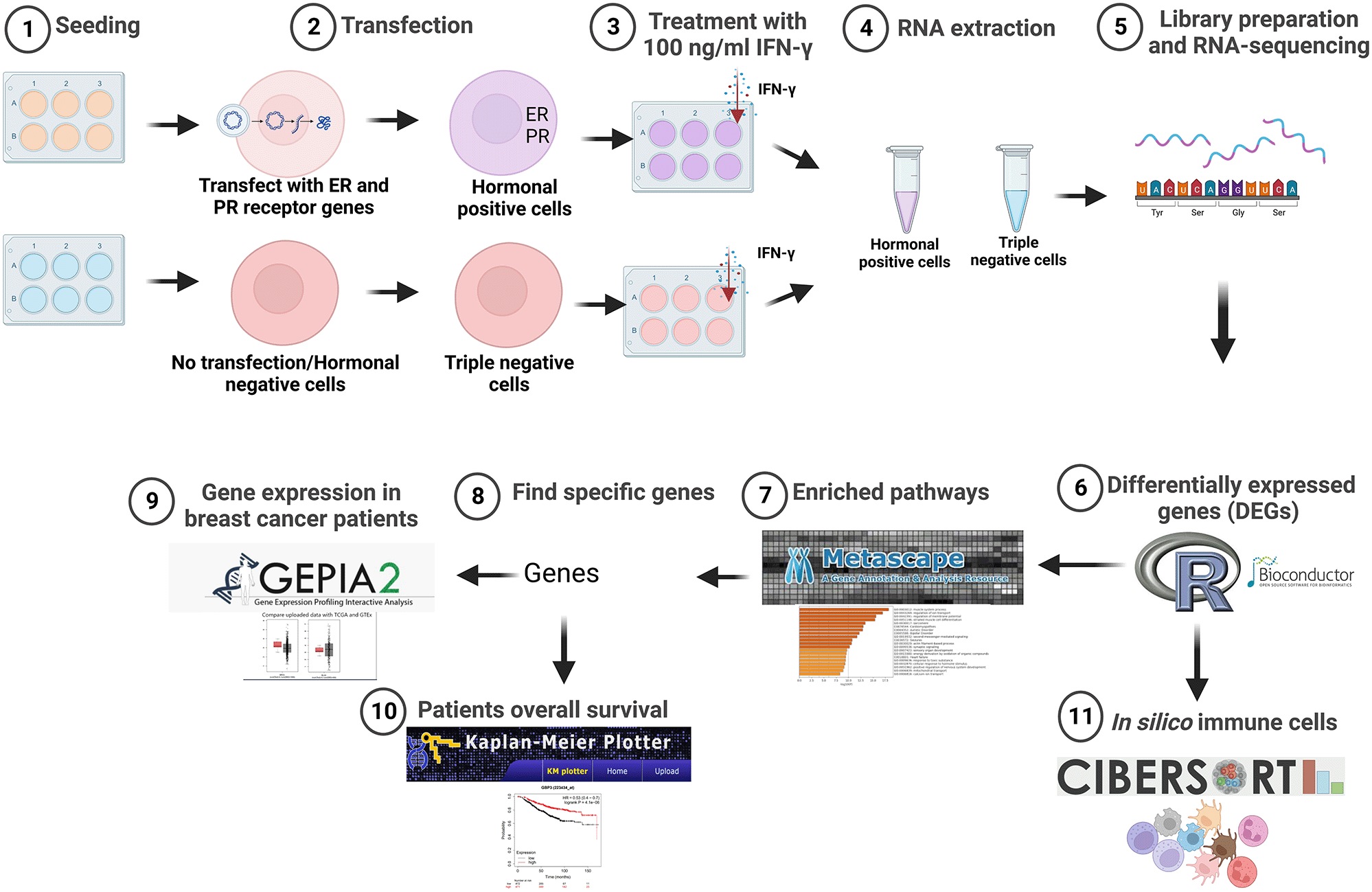
Created with BioRender.com. IFN-γ, interferon gamma; ER, estrogen receptor; PR, progesterone receptor.
Human BC cells MDA-MB-231(ER-, PR-, HER2-) were obtained from American Type Culture Collection (Manassas, VA, USA). Cells were cultured in RPMI-1640 media (Sigma-Aldrich, Cat # R8758, Germany) supplemented with 10% fetal bovine serum (Sigma-Aldrich, Cat # F9665, Germany) and 1% penicillin-streptomycin (Sigma-Aldrich, St. Louis, MO, USA) and incubated at 37°C and 5% CO2 in a humidified incubator.
MDA-MB-231 cells were seeded one day before the transfection experiment as 2×105 cells in 2 ml per well of a 6-well plate. On the day of transfection, MDA-MB-231 cells were co-transfected with 1 μg pcDNA-PR (RRID:Addgene_89130) and pEGFP-CI-ER alpha (RRID:Addgene_28230) expression plasmid construct for PR and ER, respectively, using ViaFect™ Transfection reagent (Promega; E4982). Briefly, the plasmids were diluted and mixed in 200 μl serum-free Opti-MEM™ medium (Gibco, Cat# 31985-070), then 6 μl ViaFect™ Transfection reagent was added with the ratio of plasmid DNA in μg to ViaFect Reagent in μl was 1:3. The ViaFect™ Transfection complex (Reagent:DNA mixture) was incubated for 10 minutes at room temperature and added to the well of a 6-well plate containing 2 ml of cells in growth medium drop by drop. Two biological replicates were done. Negative control cells were treated with ViaFect reagents only without adding any plasmids. After 24 hours, transfected and non-transfected cells were collected by trypsinization for measuring transfection efficiency.
RNA was extracted from cell pellets using RNA extraction kit (Norgen Biotek Corp, Cat# 47700, Thorold, Canada). Briefly, 300 μL lysis buffer was added to the pellet to lyse cells, then 300 μL of 70% ethanol was added and mixed by pipetting for 15 seconds, total lysates were transferred onto a separating column and centrifuged for 2 minutes at 3,500 x g, 400 μL wash solution was added to the column and centrifuged at 3,500 x g for 1 minutes, then wash solution was added again and centrifuged for another 2 minutes. Lastly, 50 μL Elution solution was added to elute the RNA and centrifuged for 2 minutes at 200 x g for 1 minute. cDNA was synthesized from total RNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Cat # 4368813). A total volume of 20 μL is synthesized from 1,000 ng of total RNA template. RNA samples were mixed with Nuclease-free H2O for up to 10 μL (containing 1,000 ng), the component of the cDNA kit were added together for up to 10 μL, then mixed with the RNA for a total 20 μL and loaded into thermal cycler Eppendorf™ Mastercycler® nexus gradient (Eppendorf, Cat #6331000041, RRID:SCR_023266) with thermal conditions as following: 10 minutes at 25°C, 120 minutes at 37°C, and 5 minutes at 85°C.
Quantitative PCR analysis was carried out to quantify the mRNA levels of different genes, and expression of these candidate genes was normalized against the internal control GAPDH of the same sample. For mRNA quantification, the relative expression of these genes was obtained using the 2-ΔΔCt method.17 All primers were reconstituted with Tris-EDTA (TE) buffer to a concentration of 100 μM. Then, forward and reverse primers were diluted at 1:10 in nuclease free water at the time of the experiment, and 500 ng/μL cDNA was prepared by mixing cDNA sample with nuclease free water for a total volume of 20 μL. Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher Scientific, Cat # K0223) was used for qPCR set up, 9.5 μL of the mixture was mixed with 0.5 μL cDNA for a total volume of 10 μL. The reaction tubes were loaded into the QuantStudio 5 Real-Time PCR (Applied Biosystems, Cat # A28567, RRID:SCR_020240). Thermal cycling conditions were as follows: Optimal UDG pre-treatment one cycle for 2 minutes at 50°C, then initial denaturation for one cycle for 10 minutes at 95°C, followed by 40 cycles of denaturation at 95°C for 15 seconds, annealing for 30 seconds at 60°C, then extension at 72°C for 30 seconds. The primer sequences for PR, ER, and GAPDH were as follows: PR Forward: CACAAAACCTGACACCTCCA, Reverse: TTCGAAAACCTGGCAATGAT; ESR1 Forward: GTGCCTGGCTAGAGATCCTG, Reverse: ATTTTCCCTGGTTCCTGTCC; GAPDH Forward: CCAGGTGGTCTCCTCTGACT, Reverse: ACATACCAGGAAATGAGCTT. Results were analyzed with GraphPad Prism 9 (RRID:SCR_002798). The statistical tests can be carried out using freely available alternative software such as the R programming language and JAMOVI.
After 24 hours transfection, the cells were collected and disrupted by sonication (at 40% amplitude for 10 seconds) in M-PER Mammalian protein extraction reagent (Thermo Scientific, Cat #78501) containing 1% protease inhibitor cocktail (Sigma-Aldrich Cat # P2714) and 1 mM DTT (Sigma-Aldrich Cat # 43815), which were added prior to use. Protein concentrations were determined by the Bio-Rad Protein Assay Dye Reagent Concentrate (Bio-Rad Cat # 5000006, USA). Proteins were mixed with NuPAGE LDS Sample Buffer (2x) (Life Technologies Cat # NP0008) at a 1:1 ratio and heated for 5 minutes at 95°C. Equal amounts of proteins were loaded (60 μg) then separated by 10% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane (Bio-Rad, Cat # 1620112, Germany). Membranes were blocked with 5% non-fat milk for 1 hour at room temperature, then incubated with primary antibodies overnight at 4°C. The next day, membranes were washed three times with TBST, and secondary antibodies were added for 1 hour at room temperature. The blots were developed using Clarity Western ECL substrate (Bio-Rad, Cat # 170-5061, USA). The Bio Rad ChemiDoc™ MP Imaging System (Bio-Rad, Cat # 12003154, RRID:SCR_019037) was used for chemiluminescence imaging, and images were analyzed with Image Lab software (version 6.1, Bio-Rad, RRID:SCR_014210). Western blotting images were cropped to separate between PR or ER transfection, and to remove unused wells. The antibodies used in the study were: Anti-ERα (Cell Signaling Technology, Rabbit monoclonal, Cat # 8644S, RRID:AB_2617128, dilution 1:1,000), Anti-PR A/B (Cell Signaling Technology, rabbit monoclonal, Cat # 8757S, RRID:AB_2797144, dilution 1:1,000), Anti-β-actin (Sigma-Aldrich, mouse monoclonal, Cat # A5441, RRID:AB_476744, dilution 1:3,000), Anti-rabbit IgG, HRP-linked (Cell Signaling Technology, goat anti-rabbit, Cat # 7074S, RRID:AB_2099233, dilution 1:2,000) and anti-mouse IgG, HRP-linked (Cell Signaling Technology, horse anti-mouse, Cat # 7076S, RRID:AB_330924, dilution 1:2,000).
The triple negative MDA-MB-231 cells and the transfected MDA-MB-231 cells were seeded at a density of 3x105. At 80% confluency, the cells were washed with PBS (Sigma-Aldrich, Cat # D8537, Germany) and cultured with recombinant human IFN-γ (R&D Systems, Cat # 285-IF-100, Minneapolis, MN, USA) at a concentration of 100 ng/ml. The cells were then incubated at 37°C and 5% CO2 in NuAire incubator NU-5700 (RRID:SCR_023278) for 24 hours.
Genomic DNA removal was ensured by treating the RNA (from duplicate samples of IFN-γ treated and untreated hormone-positive and TNBC MDA-MB-231 cells) with Turbo DNase (Thermo Fisher Scientific, Cat # AM1907, USA). Next, whole transcriptome sequencing was performed using targeted RNA-Seq with Ion AmpliSeq™ whole transcriptome human gene expression kit (Thermo Fisher Scientific, Cat # A26325, Massachusetts, USA). Briefly, cDNA was synthesized using a SuperScript™ VILO™ cDNA Synthesis kit (Thermo Fisher Scientific, Cat # 11754050, USA) and amplified using Ion AmpliSeq gene expression core panel primers. Amplified products were proceeded for enzymatic shearing to get amplicons of ~200 bp, then ligated with the adapter and the unique barcodes. Next, the constructed library was purified using Agencourt AMPure XP Beads (Beckman Coulter, Cat # A63881, Indianapolis, USA), quantified using an Ion Library TaqMan™ Quantitation Kit (Applied Biosystems, Cat # 4468802, Waltham, USA), further diluted to 100 pM, pooled equally, and amplified using emulsion PCR on Ion OneTouch™ 2 instrument (OT2) (Thermo Fisher, RRID:SCR_023289, Cat # 4474778, USA) and the enrichment was performed on Ion OneTouch™ ES (Thermo Fisher, Cat # 4469495, RRID:SCR_023290, USA).18 Finally, RNA-sequencing was performed on the Ion S5 XL Semiconductor sequencer using the Ion 540-Chip (Life Technologies, Cat # A27765, California, USA).
RNA-Seq data were extracted and proceeded using an in-house pipeline.19 The raw sequencing reads were aligned to the GRCh37/hg19 Human reference genome. The differentially expressed genes (DEGs) were identified between IFN-γ treated and IFN-γ untreated ER-PR transfected and un-transfected MDA-MB-231 cells using the DESeq R/Bioconductor package (Version 4.1.0, RRID:SCR_006442).20 The DEGs were selected based on a log2 fold change ≥ 2 or ≤ -2 and p < 0.05. Shared and unshared DEGs were determined using Venn diagram plots.21 Volcano plots were generated using Enhanced Volcano package in R.22 Biorender (RRID:SCR_018361) was used to put volcano plots and Venn diagrams together, and to show the flowchart summary, Chemix is a freely available alternative software that can be used instead of Biorender.
Functional enrichment analysis was performed for each DEGs list using Gene Ontology (GO) (RRID:SCR_002811)23,24 and Kyoto Encyclopedia of Genes and Genomes (KEGG) (RRID:SCR_012773) databases.25–27 GO and KEGG terms with a p < 0.05 were mapped as significantly enriched in the DEGs list. The functional clustering and pathways for the identified genes were performed using the Metascape tool (version v3.5.20230101, RRID:SCR_016620).28 In addition, a digital cytometry CIBERSORT (RRID:SCR_016955)29 was applied to the upregulated DEGs genes in IFN-γ treated compared to untreated cells in order to estimate immune cell type abundance in each MDA-MB-231 cell group. The in-silico cell fraction prediction was performed using LM22 reference gene lists for BC.30
The prognostic impact of top candidate genes was assessed in patients with BC using the Kaplan-Meier Plotter (RRID:SCR_018753),31 which is an online database that can assess the effect of gene expression on the overall survival (OS) of a large cohort of patients with BC.32 Overall, 1,880 cases of patients with BC were used in the Kaplan-Meier plotter analysis from Gene Expression Omnibus (GEO) (RRID:SCR_005012),33 European Genome phenome Archive (EGA) (RRID:SCR_004944),34 and The Cancer Genome Atlas (TCGA) (RRID:SCR_003193).35 The Expression Analysis module of the Gene Expression Profiling Interactive Analysis, version 2 (GEPIA2) (RRID:SCR_018294)36 web server was used to obtain the expression differences of IL-6 and CCL5 tumors and adjacent normal tissues with p-value cutoff = 0.01, log2FC (fold change) cutoff = 1.
The Kaplan-Meier survival plots with number at risk, hazard ratio (HR), 95% confidence intervals (CI) and log-rank P-values were obtained using the Kaplan-Meier plotter website. For validation, statistical differences between two groups were analyzed using unpaired Student’s t-test. p < 0.05 was considered to be statistically significant.
The differential gene expression analysis showed different transcriptional profiling between hormone receptor-positive and TNBC MDA-MB-231 cells after treatment with IFN-γ. Following transcriptomic data normalization and filtering, a total of 191 and 512 genes were differentially expressed (FC > 2 and p < 0.05) in ER- and PR-positive BC cells after treatment with IFN-γ cytokine compared with untreated cells and in the TNBC cells after treatment with IFN-γ cytokine compared with untreated cells, respectively (Table 1).37
BC, breast cancer; DEGs, differentially expressed genes; ER, estrogen receptor; PR, progesterone receptor.
| Comparison | MDA-MB231/ER-PR Treated vs. ER-PR-Untreated | MDA-MB231/Treated vs. control |
|---|---|---|
| All DEGs (p < 0.05) | 191 | 512 |
| Upregulated genes (p < 0.05) | 159 | 146 |
| Downregulated genes (p < 0.05) | 32 | 366 |
DEGs were investigated with regard to the expression of the hormone receptor genes (ER and PR). Most genes were found to be unique to the cell type when treated with IFN-γ. Only 29 genes were found to be commonly differentially expressed in both types of cells (Figure 2).
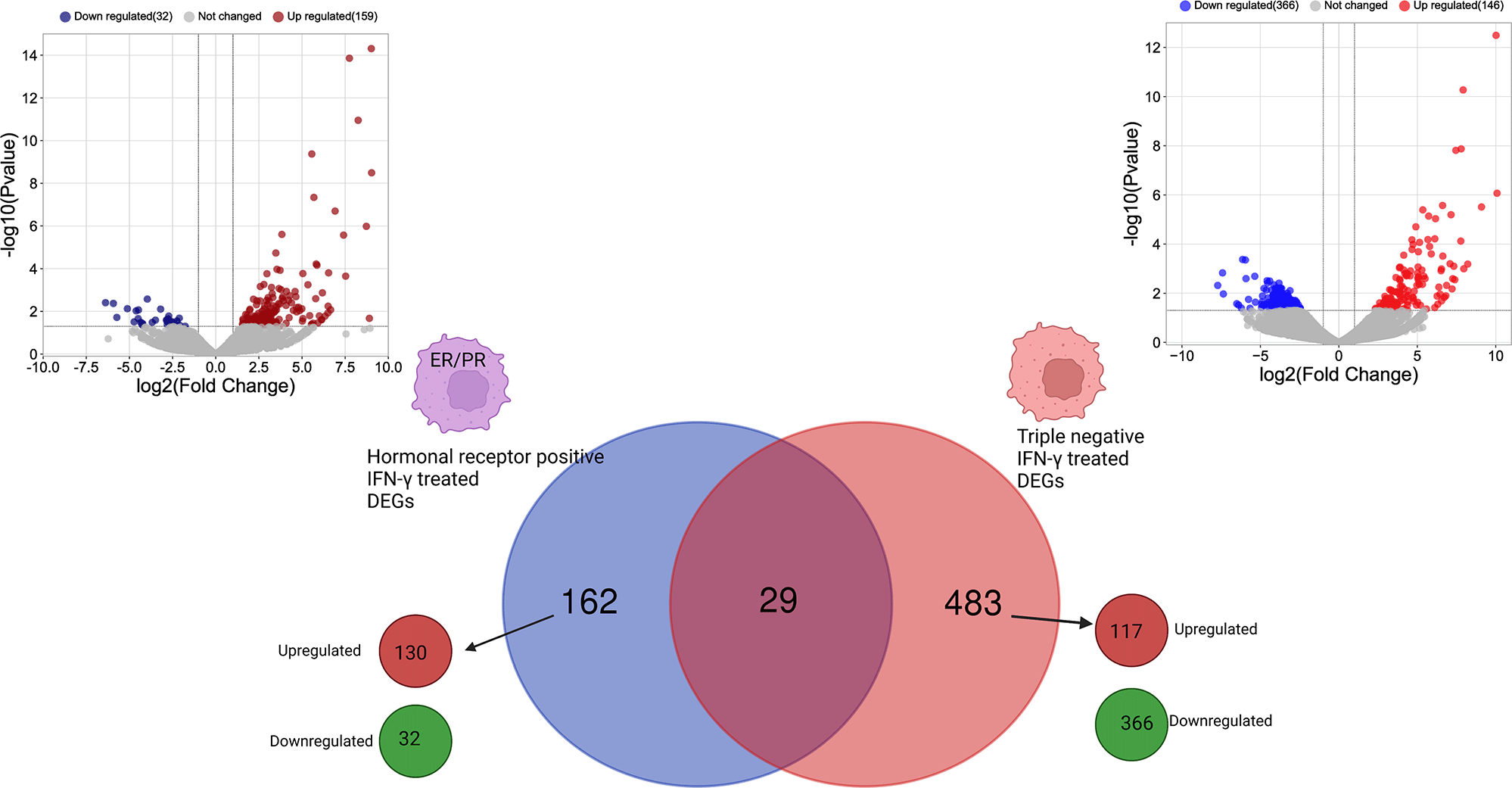
Created with BioRender.com. IFN-γ, interferon gamma; ER, estrogen receptor; PR, progesterone receptor; DEGs, differentially expressed genes.
Functional enrichment pathways indicate enhancement of some pathways shared between both types of BC cells regardless of the ER and PR expression. Among these pathways are “hsa04612: antigen processing and presenting”, “GO:0006954: inflammatory response”, “GO:0042157: lipoprotein metabolic process”, and “GO:0031347: regulation of defense response” pathways (Figure 3).
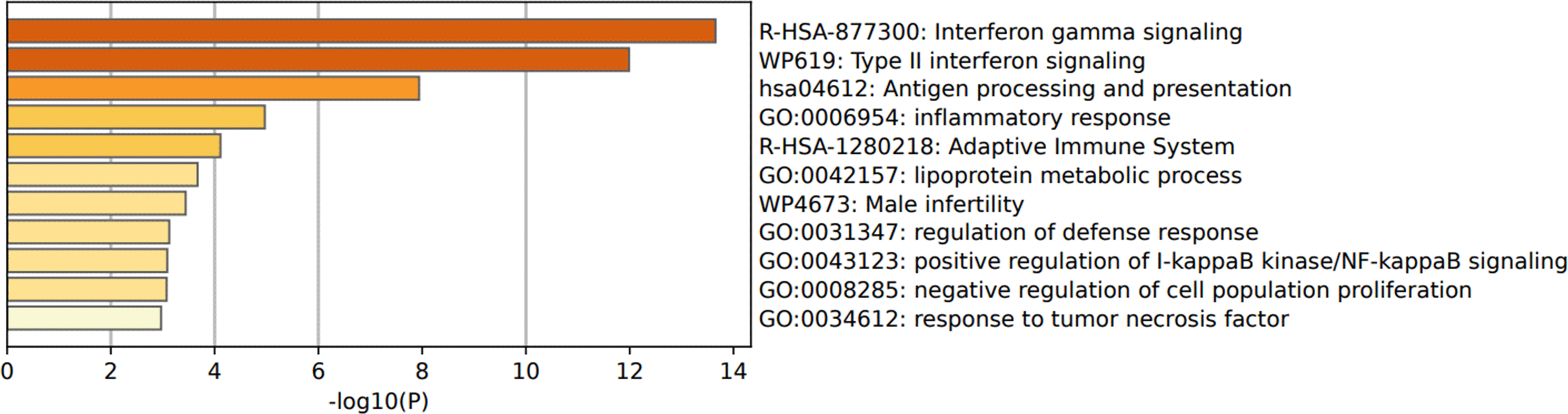
IFN-γ, interferon gamma; DEGs, differentially expressed genes.
IFN-γ induces a modulation of 162 unique DEGs in ER- and PR-positive cells, among them, 130 genes were upregulated, and 32 genes were downregulated. While in the TNBC cells, 483 unique DEGs were identified including 117 upregulated genes and 366 downregulated genes (Figure 2).
Functional clustering and pathways analysis of the upregulated genes in the hormone-positive and TNBC MDA-MB-231 cells after treatment with IFN-γ revealed differences in genes expression in both types of cells. Both cell types showed a response to IFN-γ by upregulating some functional pathways. These pathways are: “R-HAS-877300: Interferon gamma signaling”, and “R-HAS-1280218: Adaptive immune system” were among the enriched pathways upregulated in ER and PR positive cells. Similarly, the pathways “GO:0071346: cellular response to interferon-gamma”, and “GO:0045824: negative regulation of innate immune response” were among the pathways enriched in TNBC cells (Figure 4).
Notably, the genes involved in the immune system pathway are different between the two cell types, as shown in Table 2. The ER and PR positive cells showed upregulated genes that are involved in the “R-HSA-1280218: adaptive immune system” pathway, including B2M, CD74, CTSS, HLA-B, HLA-DPA1, HLA-DRB1, HLA-E, ICAM1, PPP2R1A, PSMB8, PSMB10, PSME2, TRIM21, ERAP1, SPSB1 and ZNRF1 (Figure 4A, Table 3). While the TNBC cells showed that upregulated genes including CEACAM1, LGALS9, NMI, ISG15, TRAFD1, and SLAMF8 modulate the “GO:0045824: negative regulation of innate immune response” (Figure 4B, Table 3).
BC, breast cancer.
TNBC, triple negative breast cancer; GO, Gene Ontology.
Based on the functional clustering and pathways analysis of the upregulated genes, the ER- and PR-positive cells showed the activation of B2M, GBP3, HLA-B, HLA-DPA1, HLA-DRB1, HLA-E, ICAM1, OAS3, PML, TRIM21, and STAT1 as significantly related genes to “R-HSA-877300: IFN-γ signaling”. The survival analysis using gene expression data on patients with BC was used to verify the prognostic relationship between the aforementioned genes and BC. All genes involved were investigated for OS in BC (Extended data37). Only the genes that showed a significant association with BC prognosis were reported (p-value < 0.05; Figure 5A-D). The highly expressed genes like GBP3 (HR, 0.52; 95% CI, 0.39–0.69; P= 3.0e-6; Figure 5A), HLA-DPA1 (HR, 0.74; 95% CI, 0.61–0.9; P= 0.0024; Figure 5B), HLA-DRB1 (HR, 0.76; 95% CI, 0.63–0.92; P=0.0049; Figure 5C), and HLA-E (HR, 0.75; 95% CI, 0.62–0.91; P=0.0041; Figure 5D) were significantly associated with improved OS in patients with BC.
Likewise, the TNBC cells showed the significant activation of the following genes CCL8, IFI6, ISG15, LGALS9, SP100, CLDN1, and NMI in the”GO:0071346: cellular response to IFN-γ”. Importantly, Kaplan-Meier curve analysis of these genes and the BC patients’ survival analysis showed that the highly expressed genes of IFI6 (HR, 1.29; 95% CI, 1.06–1.56; P= 0.0096; Figure 6A) and ISG15 (HR, 1.23; 95% CI, 1.02-1.49; P= 0.034; Figure 6B) were significantly associated with poor OS in patients with BC. The remaining genes were not significantly associated with the prognosis of BC.
To understand the role of the genes involved in functional pathways, we explored the genes that most frequently appear in functional pathways. The top five genes of each cell type were selected, IL-6, HLA-DRB1, HLA-E, PSMB10, B2M most frequently appeared in hormone receptor-positive cells pathways, and CCL5, JAK2, LGALS9, TLR3, CEBPB most frequently appeared in TNBC cells pathways. IL-6 was shown to be involved in 160 functional pathways in the hormone receptor-positive cells. CCL5 was shown to be involved in 135 functional pathways of the TNBC cells (Figure 7A and B). These top frequently appearing genes were further assessed across various cancer types. The GEPIA2 database explored IL-6 and CCL5 expression in various cancer types, and BC showed significant differential expression between normal and tumor tissues. IL-6 showed high expression in normal tissue (n= 291) compared with adjacent breast tumor tissue (n= 1,085), while CCL5 gene showed higher expression in breast tumor tissue (n= 1,085) compared with adjacent normal tissues (n= 291) (Figure 7C-F).
Next, we investigated the downregulated genes in both cell types. The downregulated genes of hormone receptor-positive cells were only associated with the “GO:0043414: macromolecule methylation” and “GO:0006091: generation of precursor metabolites and energy” pathways (Figure 8A). While the downregulated genes in TNBC cells were mainly enriched to cell organization and synthesis, including “GO:0000278: mitotic cell cycle”, “R-HSA-1852241: organelle biogenesis and maintenance”, and “GO:0007005: mitochondrion organization” pathways (Figure 8B).
Since BC cells treated with IFN-γ showed differences in functional clustering pathways, we wanted to explore the effect of this cytokine on the abundance of related immune cells. An in-silico analysis of the differential expressed genes of each cell type was performed using CIBERSORT to find the expected abundance of the immune cells. As expected, immune related gene expression in hormone receptor-positive cells was significantly different from that in TNBC cells (Figure 9). In hormone receptor-positive cells, a distinct increase was detected in the adaptive immune gene expression of CD4 naïve T cell (19.5%) and B cells (3.7%), in addition to an increase in neutrophils (32.5%), NK cells (15.2%), M2 (13.4%) eosinophils (10.5%), and an obvious activation on monocytes (4.8%) (Figure 9). However, TNBC cell lines showed an increase mainly in M1 (21.5%), and M2 (28.8%) (Figure 9). Moreover, the TNBC cells do not show the major innate immune cells related genes e.g., dendritic cells (DCs), eosinophils, neutrophils, NK cells, TCR-γδ+ T cells, activated mast cells, and M0 (Figure 9).
IFN-γ is known to be highly released from different types of cells, such as the innate immune cells NK cells, and cells of the adaptive immune response, such as CD8+ and CD4+ T cells.38,39 Previously, IFN-γ was believed to have antitumor activity by modulating immune cells in the TME besides its direct pro-apoptotic effect on cancer cells.40 However, recent studies have shown that IFN-γ is a cytokine that has both protumor and antitumor activities.41 It may contribute to tumor progression and metastasis in different ways such as increasing PD-L1 expression on the tumor cell surface leading to tumor escape immune detection.42,43
The presence or absence of ER/PR is critical to BC cells and forms the basis for its classification.44 MDA-MB231 cells transfected with ER/PR, provide an alternative in vitro model to the MCF7 ER/PR cell line. This transfection allows us to evaluate the specific effect of PR and ER without interference with other biological components.45,46
In the present study, IFN-γ induced a significant change between the transcriptome of hormone receptor-positive and TNBC MDA-MB-231 cells. It has been reported that the cytokine network, such as IL-6, IL-8, IL-10, IL-17, IL-1β, TNF-α, IFN-γ, and MCP-1, has distinctive functional features on the status of the immune system, and the patient’s prognosis depends on the BC subtype.47,48
IFN-γ induced upregulation in DEGs and enhanced the IFN-γ signaling pathway and adaptive immune system pathway in ER- and PR-positive cells. Our study revealed the upregulation of the adaptive immune pathways, which have a crucial role in normal tissue development and breast tumor suppression. The adaptive immune cells, mainly T lymphocytes, are reduced in number in BC and have been correlated with tumor rejecting capacity.49,50 Immune cells within TME communicate with each other and with tumor cells in order to control tumor growth,51 and an increase in immune cell infiltration is associated with a favorable prognosis of BC.10 This explores the role of IFN-γ in the induction and regulation of adaptive immune responses, which may implicate tumor regression and death in the ER/PR-positive BC cells.
By contrast, the functional clustering and pathway of the TNBC cells showed a negative regulation of the innate immune response in the IFN-γ treated BC cells. Immunosurveillance is a term referring to innate immune cells’ recognition and rejection of cancer cells at an early stage of tumor initiation. The innate immune cells, mainly NK cells, control the abnormal growth of cancer cells by secreting various cytokines such as IFN-γ.52,53 Our finding indicates that IFN-γ treated TNBC cells attained an immune regulatory phenotype in which negative regulation of the innate immune system was highly enhanced, and this might further promote tumor progression and survival in this type of cell.
The IFN-γ signaling pathway is highly associated with the OS of patients with BC. The upregulation in GBP3, HLA-DPA1, HLA-DRB1, HLA-E transcriptome predicted a good prognosis. GBP3 participates in host defense against pathogens and transformed cells, and induces apoptosis in cancer cells other than normal cells.54 The expression of HLA, composed of an α chain (DRA) and a β chain (DRB), is highly associated with the activation of antitumor immunity through interactions with T lymphocyte receptors and produces an active antitumor immune response thereby improving the survival and prognosis of patients with cancer.55,56 In various healthy tissues, expression of nonclassical HLA class I molecules (HLA-E and HLA-G), play an essential role in immune surveillance by NK cells.56 Elevated levels of these genes is associated with good prognosis in patients with cancer. Upregulation of these genes by IFN-γ treatment may have a protective role in ER- and PR-positive cells by inducing tumor cell death and activating immune defense.
The TNBC cell-associated genes with IFN-γ pathway (IFI6, ISG15) showed a negative association with the OS of patients with BC. IFI6, also known as ISG 6-16, is one of the IFN-γ-induced genes that has been reported to have antiapoptotic effects on some types of cancer, including BC.57 ISG15 protein conjugates with many different cellular proteins involved in regulation of protein turnover that might also be associated with tumorigenesis in cancers originating from other tissue types.58,59 Our findings suggest that upregulating these genes in TNBC cells by IFN-γ treatment, may exacerbate tumor progression by upregulating oncogenic genes. We suggest that the role of IFN-γ in patients with TNBC should be considered when customizing treatments. Our analysis of the top frequently appearing genes in functional pathways revealed that IL-6 and CCL5 mostly appeared in hormone-positive and TNBC cells, respectively. IL-6 is a cytokine that is known to be involved in the regulation of tissue repair besides its antitumor cell immune response in TME.60,61 The chemokine CCL5, also called Regulated upon Activation, Normal T Cell Expressed and Presumably Secreted (RANTES), has been associated with cell proliferation, migration, angiogenesis, metastasis, and survival.62,63 Showing IL-6 gene expression in hormonal receptor positive cells, and CCL5 gene expression in TNBC cells may further confirm the opposing role of IFN-γ on hormone receptor and TNBC cells.
In the present study, IFN-γ induced a significant modulation in immune cell related genes of both BC cell types. The increase in neutrophils, eosinophils, T cell, NK cells, monocyte, and B cells suggest that hormone receptor-positive cells enhance their immune defense against tumor cells when treated with IFN-γ. M1 macrophages display the polarized state that characterizes the antitumor activity, while M2 macrophage display the state that directs to tumor promotion.64,65 M1/M2 ratio defines the survival outcome and destiny of the tumor cells.65,66 By comparison, our results for the TNBC cells showed higher increases in M2 than in M1. The M1/M2 ratio indicates immune suppression and poor tumor prognosis. In addition, the missing gene expression of the important innate immune cells, DC, eosinophils, neutrophils, NK cells, T cell γδ, and activated mast cells contributes to immune escape and tumor progression of this type of cancer, and these results support our earlier findings of the negative regulation of the innate immune pathway.
In this study, we applied several bioinformatics analyses on whole transcriptomics data on triple negative and hormone receptor-positive cell lines and validated it using patient data obtained from publicly available sources to explore the effect of ER and PR on tumor progression in the presence of IFN-γ. The results showed that genes upregulated in hormone receptor-positive cells (GBP3, HLA-DPA1, HLA-DRB1, HLA-E, IL6) and TNBC cells (IFI6, ISG15, CCL5) had roles in immune modulation and tumor progression as well as patient prognosis. Upregulated genes in IFN-γ treated hormone receptor-positive BC push tumor cells toward immune activation and patients’ survival, while upregulated genes in IFN-γ treated TNBC induce negative immune responses and thus decrease patients’ overall survival. These results may provide novel insights into the IFN-γ molecular mechanism in BC with respect to the ER/PR/HER2 status and may form the basis to improve personalized therapy for patients with BC with better results and minimal side effects.
Figshare: Impact of estrogen and progesterone hormone receptors on the progression of interferon-γ sensitized breast cancer cells. https://doi.org/10.6084/m9.figshare.21814527. 37
This project contains the following underlying data:
• Data file 1 RawData Transcriptome IFNγ BC (1).xlsx (Whole transcriptomic raw for ER/PR transfected and un-transfected MDA-MB-231 cell line treated with 100 ng/ml IFN-γ)
• Data file 2 Processed IFNγ BC data (1).xlsx (ER/PR transfected and un-transfected MDA-MB-231 cell line treated with 100 ng/ml IFN-γ)
• Data file 4 ER-PR raw data PCR results.xlsx
• Data file 5 ER-PR raw data western blots.docx
• Data file 6 ER-PR raw data western blots.zip
This project contains the following extended data:
• Data file 3 Extended data.docx (Validation of transfection, overall survival in breast cancer patients, CIBERSORT table).
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
The authors would like to thank staff at the University of Sharjah Tissue Bank for their support.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Expertise: breast cancer models, hormone receptors. Not an expert in the bioinformatic analysis
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Yes
If applicable, is the statistical analysis and its interpretation appropriate?
Partly
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: sc RNA-seq
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Yes
Are all the source data underlying the results available to ensure full reproducibility?
Partly
Are the conclusions drawn adequately supported by the results?
Yes
Competing Interests: I collaborated with Dr Rifat Hamoudi more than 5 years ago when he was a staff member at the University College London. We have not published any scientific manuscripts as co-authors in the last 3 years.
Reviewer Expertise: Cancer, Signaling pathways in cancer, Biomarker discovery in epithelial cancers
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 1 08 Sep 23 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)