Keywords
Neohylomys hainanensis, mitochondrial genome, phylogeny, Erinaceidae
This article is included in the Cell & Molecular Biology gateway.
This article is included in the Genomics and Genetics gateway.
The Hainan Gymnure Neohylomys hainanensis is a small-size mammal which occurs in Hainan, China and north Vietnam. Here, we report the complete mitochondrial genome of N. hainanensis. The whole mitochondrial genome is 17,337 bp, and contains 13 protein-coding genes, 22 tRNAs, 2 rRNAs and one control region. The base composition of the N. hainanensis total mitogenome is: 33.4% A, 12.2% G, 33.1% T, and C 21.3%, with an A + T content of 66.5%. The K2P genetic distance analysis supports current taxonomy that places N. hainanensis, Hylomys suillus and Neotetracus sinensis in different genera. Phylogenetic analysis suggests that N . hainanensis is closely related to Neotetracus sinensis based on the complete mitochondrial genome sequences. The mitogenomic data will contribute to molecular phylogenetics and conservation genetics of the species.
Neohylomys hainanensis, mitochondrial genome, phylogeny, Erinaceidae
The article has been revised following two reviewers' comments and suggestions. The readability of the article has been improved.
See the authors' detailed response to the review by Michael Allen
See the authors' detailed response to the review by Perinçek Seçkinozan Şeker
The Hainan Gymnure (Neohylomys hainanensis Shaw and Wang, 1959) is a small mammal distributed in Hainan, China and north Vietnam, where it inhabits tropical and subtropical forests (Liu and Wu 2019; Shaw et al. 1966; Smith et al. 2009; Abramov et al. 2018). In China, eight species are currently recognized within the family Erinaceidae (Smith et al. 2009). However, the phylogenetic relationships of three species (N. hainanensis, Hylomys suillus and Neotetracus sinensis) within Erinaceidae have been debated (Cobert 1988; Gould 1995; Grenyer and Purvis 2003; He et al. 2012). Previous studies (Cobert 1988; Gould 1995; Grenyer and Purvis 2003) indicated sister relationships of N. hainanensis and N. sinensis based on morphological data. However, He et al. (2012) showed that N. sinensis clustered with H. suillus rather than N. hainanensis based on combined data of mitochondrial genes (CYTB, ND2 and 12S). Complete mitochondrial genome sequences are highly informative markers for resolving phylogenetic relationships among taxa (Kumazawa 2007; Shen et al. 2010; Yue et al. 2015). Though the complete mitochondrial genome of N. hainanensis is available in GenBank (MW429379), we sequenced the mitochondrial genome using our sample based on primer walking method to confirm the accuracy of the data. Additionally, we investigated phylogenetic relationships of N. hainanensis among family Erinaceidae in an attempt to resolve the current phylogenetic disputes.
The animal was captured using a mouse cage trap at Yinggeling Mountains (109.289°E, 18.878°N), Hainan province, China in May, 2022. The animal was euthanized in a 10 L euthanasia chamber, which was gradually filled with 99% purity of CO2 within 2-3 minutes.
A total of 2 g of femoral muscle sample was clipped from the specimen using surgical scissors, and the total genomic DNA was extracted from the muscle tissue using a standard phenol-chloroform extraction protocol (Sambrook et al. 1989), which contained three main procedures: 1) Sodium dodecylsulfate (SDS) and proteinase K were used for the enzymatic digestion of proteins; 2) A mixture of phenol: chloroform:isoamyl alcohol (25:24:1) was then added to promote the partitioning of lipids into the organic phase; 3) The DNA was rinsed using analytical alcohol of different solubility and then otained the purified DNA for PCR. Seven primer combinations were used for the generation of PCR products (see Extended data (Tu 2023)). PCR amplifications of mitochondrial genome of N. hainanensis were listed as follows: PCR amplifications were carried out in 25 uL reaction volumes containing 1×EX Taq buffer (Mg2+ Free; TaKaRaBiotech, Dalian, China), 2.5 mM MgCl2, 0.1-0.4 mM dNTP, 0.4 uM each primer, 0.04 u/uL EX Taq polymerase, and ~5 ng/uL total genomic DNA as template. PCR protocol was set as follows: an initial denaturation at 94°C for 4 min; 35 cycles of amplification as the main procedure: a denaturation at 94°C for 45 s, an annealing at 50-60°C for 50 s, an elongation at 72°C for 2-3 min, and a final extension at 72°C for 4 min. All PCR products were examined through electrophoresing on a 1.0% agarose gel and then directly sequenced using an ABI 3730xl sequencer. The complete mitochondrial genome sequence was then assembled and manually refined utilizing SeqMan (DNASTAR 7.1.0) (Swindell and Plasterer 1997) following analysis of chromatogram files. The boundaries of protein-coding genes were predicted via comparison with homologs using MEGA 6.0 (Tamura et al. 2013). The transfer RNA (tRNA) genes were identified using tRNAscan-SE 2.0 (Lowe and Chan 2016).
To better understand the phylogentic position of N. hainanensis within Eulipotyphla, we constructed a phylogenetic tree by using neighbor-joining (NJ) method implemented in MEGA6.0 (Tamura et al. 2013) based on 13 complete mitochondrial genome sequences of Eulipotyphla using Kimura 2-parameters (K2P) model.
The whole mitochondrial genome of N. hainanensis is 17,337 bp in length (GenBank Accession number ON054206.2 (Tu and Hou 2022)) and contains 13 protein-coding genes (PCGs), 22 tRNAs genes, two rRNAs genes, and one control region. The base composition of N. hainanensis mitogenome is as follows: A 33.4%, G 12.2%, T 33.1%, and C 21.3%. Typically, an A+T rich pattern (66.5%) was observed. Comparison with a previously published mitochondrial genome sequence (MW429379) for the same species, these two sequences differ in length (17,337 bp vs 16,795 bp), which may be due to mitochondrial control region sequence variations (1,868 bp vs 1334bp). Within 37 mitochodrial genes, the ND6 gene and eight tRNAs (tRNA Gln, tRNA Ala, tRNA Asn, tRNA Cys, tRNA Tyr, tRNA Ser, tRNA Glu and tRNA Pro) were encoded on the L-strand, whereas all other genes were encoded on the H-strand. Most mitochondrial PCGs, use ATG as its start codon, ND2 and ND3 begin with ATT, and ND5 begin with ATA. Most mitochondrial PCGs contain ATG as the start codon, ND2 and ND3 begin with ATT, and ND5 begins with ATA.
A phylogenetic tree showed that two sequences of N. hainanensis shared a common evolutionary ancestry. Within subfamily Galericinae, H. suillus and the common ancestors of N. hainanensis and N. sinensis formed sister groups ( Figure 1). The results were consistent with morphological studies (Cobert 1988; Gould 1995; Grenyer and Purvis 2003). Our results support current taxonomy that places the N. hainanensis, N. sinensis and H. suillus in different genera (Smith et al. 2009; Wei et al. 2021). This mitochondrial genome sequence determined in this study will benefit future research into conservation genetics and phylogenetics for the species.
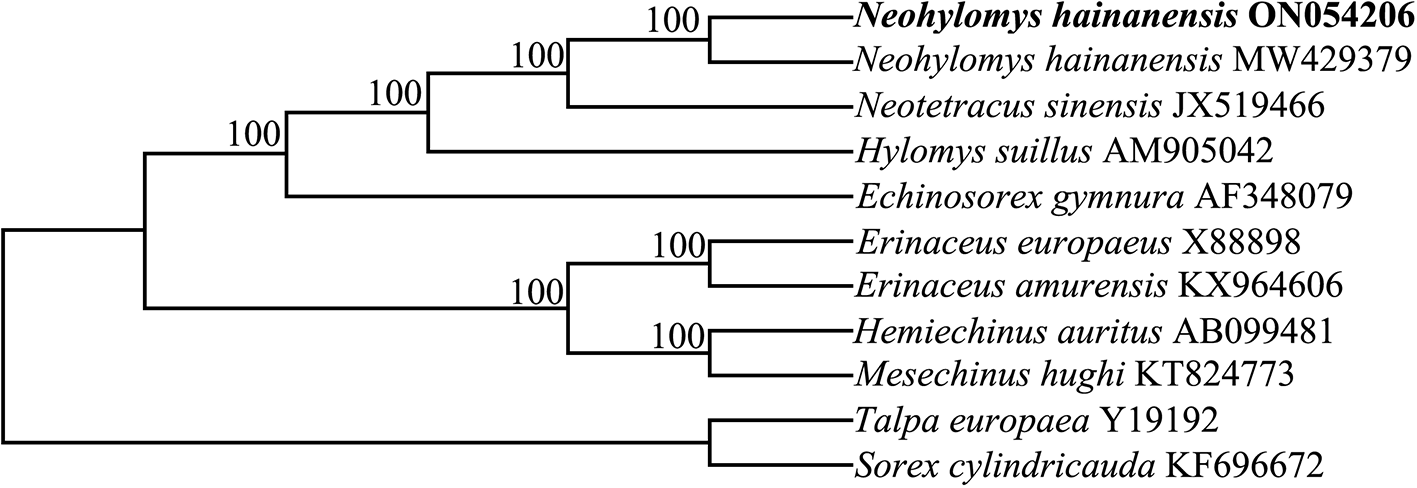
Nodal support indicated by bootstrap. The codes after the species names represent GenBank accession numbers.
This work complies with the International Union for Conservation of Nature (IUCN) policies research involving species at risk of extinction (see Guidelines for appropriate uses of IUCN Red list data version 4.0; https://www.iucnredlist.org/resources/guidelines-for-appropriate-uses-of-red-list-data ), as the species under study is an endangered species. All procedures were approved by Animal Research Ethics Committee of Hainan Provincial Education Center for Ecology and Environment, Hainan Normal University (HNECEE-2022-003) on March 6, 2022.
NCBI Nucleotide: Neohylomys hainanensis voucher HN20N051 mitochondrion, complete genome. Accession number ON054206.2, https://www.ncbi.nlm.nih.gov/nuccore/ON054206.2 (Tu and Hou 2022).
Figshare: Neohylomys hainanensis primer combinations data. https://doi.org/10.6084/m9.figshare.22180219.v1 (Tu 2023).
This project contains the following extended data:
Data are available under the terms of the Creative Commons Attribution 4.0 International license (CC-BY 4.0).
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Conservation, Environmental DNA, DNA metabarcoding, Bioinformatics
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Mammalogy, taxonomy and phylogeny
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular Systematics Zoology
Are the rationale for sequencing the genome and the species significance clearly described?
Partly
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Molecular Systematics Zoology
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Conservation, Environmental DNA, DNA metabarcoding, Bioinformatics
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 20 Oct 25 |
read | read | read |
|
Version 1 16 Mar 23 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)