Keywords
White Paper, ELIXIR Strategy, Community Roadmap, Biodiversity Networks, Biodiversity Informatics, Environmental Systems, Data Science
This article is included in the ELIXIR gateway.
This article is included in the Bioinformatics gateway.
This article is included in the Ecology and Global Change gateway.
Biodiversity loss is now recognised as one of the major challenges for humankind to address over the next few decades. Unless major actions are taken, the sixth mass extinction will lead to catastrophic effects on the Earth’s biosphere and human health and well-being. ELIXIR can help address the technical challenges of biodiversity science, through leveraging its suite of services and expertise to enable data management and analysis activities that enhance our understanding of life on Earth and facilitate biodiversity preservation and restoration. This white paper, prepared by the ELIXIR Biodiversity Community, summarises the current status and responses, and presents a set of plans, both technical and community-oriented, that should both enhance how ELIXIR Services are applied in the biodiversity field and how ELIXIR builds connections across the many other infrastructures active in this area. We discuss the areas of highest priority, how they can be implemented in cooperation with the ELIXIR Platforms, and their connections to existing ELIXIR Communities and international consortia. The article provides a preliminary blueprint for a Biodiversity Community in ELIXIR and is an appeal to identify and involve new stakeholders.
White Paper, ELIXIR Strategy, Community Roadmap, Biodiversity Networks, Biodiversity Informatics, Environmental Systems, Data Science
The article has been substantially revised to address the points raised by the three reviewers - the responses to the reviews contain also the main new texts that were added to the manuscript during revisions. This included the addition of several new references to support statements made throughout the text. It also included providing more context about ELIXIR and the role of ELIXIR Communities in general, as well as substantially extending the description of the structure and operational functioning and goals of the Biodiversity Community and a description of the Community’s Implementation Study. Four authors were added as they contributed substantially to the manuscript revisions, especially the newly added parts about the Community goals and Implementation Study.
See the authors' detailed response to the review by Abigail Benson and Stephen Formel
See the authors' detailed response to the review by Cher FY Chow
See the authors' detailed response to the review by Patrick Comer
Biological diversity—or biodiversity—refers to the variety and variability of life on Earth, encompassing genetic and species diversity at the levels of populations, communities, and ecosystems. Biodiversity reflects the ever-changing natural balance that has evolved over billions of years, sustaining communities of interdependent and interacting organisms. Those balances form the basis of a healthy Earth, including the ecosystem functions that support human well-being (i.e., ecosystem services). With growing demands on nature due to human activities, the Anthropocene is upsetting this balance and is consequently witnessing an unprecedented loss of biodiversity globally (WWF, 2022; Johnson et al., 2017). These declines pose a grave threat to humanity, the severity of which is increasingly recognised by international organisations, regional bodies, national governments, and society. The urgency to act is recognised particularly in the field of conservation biology, which has been described as a “discipline with a deadline” (Soulé & Wilcox, 1980).
Strategies to protect and restore biodiversity are wide-ranging in scope and scale, with policies and actions that require broad support to be feasible and effective e.g., goals 12-15 of the 17 Sustainable Development Goals (SDGs) adopted by the United Nations (UN, 2015). Biodiversity research aimed at building the knowledge and resources that inform management practices and policy is equally wide-ranging, often bringing together researchers from different disciplines, such as taxonomists, ecologists, evolutionary biologists, and informaticians. This is particularly true for the growing field of interdisciplinary research taking advantage of molecular sequence data, which recognises the relevance of and advantages offered by genetic and genomic data in biodiversity assessment, monitoring, conservation, and restoration (Hoban et al., 2021; Lewin et al., 2022). Connecting such molecular sequence data with biodiversity research infrastructures (see Extended Data (Waterhouse, 2023)) and resources is a critical step towards facilitating exchange of knowledge, sharing, and interoperability of large and complex datasets (Waterhouse et al., 2022).
As a European life sciences infrastructure, ELIXIR strives to coordinate bioinformatics resources from across Europe to enable researchers to access and analyse life science data, to improve the value and impact of life science research on public health, the environment, and the economy. The need for informatics solutions to address key societal challenges inspires many scientists from across the ELIXIR Nodes to increasingly engage in different aspects of biodiversity research. This stems from a natural alignment with ELIXIR’s overarching mission to support the management of public research data, integrate and coordinate life science resources, and foster the development of innovative services and technical solutions in Europe (Harrow et al., 2021). Here we present the ELIXIR Biodiversity Community, comprised of researchers from different disciplines, united by a shared recognition of the main societal and informatics challenges, as well as key scientific and organisational opportunities; how these connect with ELIXIR Platforms and other ELIXIR Communities, as well as with the wider “ecosystem” of biodiversity projects and infrastructures; and set out our roadmap for building on ELIXIR expertise to grow the ELIXIR Biodiversity Community and engage with the development of resources and infrastructures to support biodiversity research.
Biodiversity represents the variety of organisms on the planet at all taxonomic levels, a result of a long and complex evolutionary process. Biodiversity is essential for life itself, for the adaptation of populations, species, communities, and ecosystems towards rapid change in biotic and abiotic parameters, including climate change. From a human standpoint, biodiversity forms the foundation of ecosystem services that are indispensable for human well-being and a healthy planet, and has long been a source of adaptive solutions or innovations in several critical areas such as food production. Despite its importance, biodiversity has been declining at a mass-extinction-level rate (IPBES, 2019) over the last decades. The unsustainable human development model has increased pressures on biodiversity, through climate change (IPCC, 2022; Wezel et al., 2020), invasive species, habitat loss and degradation, and the depletion of natural resources (IPBES, 2019). The decline of biodiversity at this rate often creates unpredictable threats and changes to ecological oscillations, such as the increased risk of new human diseases (Frumkin & Haines, 2019), the collapse of ecosystem services, the degradation of natural resources, and the increased possibility of a global food crisis (FAO, 2019).
At the same time, scientists and naturalists do not even know what is being lost, as around 80% of biodiversity at the species and population levels remains undescribed and/or underrepresented in inventories and databases (Mora et al., 2011; Costello et al., 2013; Moura & Jetz, 2021; Bispo et al., 2022; Boekhout et al., 2022; Chimeno et al., 2022). Furthermore, most research and monitoring efforts tend to focus on a limited number of biodiversity levels or elements. While there is significant literature around biodiversity loss (e.g., a Scopus query [13.09.2022] for “biodiversity loss” returns 33,324 documents), there is a very limited effort in reviewing biodiversity using high-throughput data (Scopus query [13.09.2022] for “Biodiversity loss” AND (“omics” OR “genomics” OR “metagenomics”) returns only 1,795 documents). This clearly indicates a bias in reporting, which has repercussions on the decision-making process pertaining to biodiversity conservation efforts (Gadelha et al., 2021). This brings forward an additional challenge of shifting perspectives from narrow, low-throughput efforts towards more holistic, high-throughput initiatives, including better citizen scientist contributions towards these efforts. Humanity may miss important solutions to key problems for its survival, such as the loss of important genetic variants among wild plants, animals, and microbes for agriculture (Nic Lughadha et al., 2020) and for dealing with health issues (Marselle et al., 2021).
Following the 1992 United Nations Convention on Biological Diversity (CBD), governments and international organisations have responded to the decline of biodiversity with policies, and restoration and protection strategies. However, the initial goals of these have not been reached and biodiversity decline continues accelerating (IPBES, 2019; Turvey & Crees, 2019; WWF, 2022). For the new targets set by the post-2020 global biodiversity framework (GBF, 2023) to succeed, research is considered to be key, especially the interaction between science, society, and policy makers (Blicharska et al., 2019; Hermoso et al., 2022; Nature, 2022), with net improvements by 2050 to achieve the CBD’s vision of “living in harmony with nature by 2050”. Today, scientists recognise the important roles that genetic and genomic data can play in biodiversity discovery, assessment, monitoring, conservation, and restoration, to ensure the long-term resilience of ecosystems (Hoban et al., 2020; Gadelha et al., 2021; Segelbacher et al., 2022; Formenti et al., 2022; Theissinger et al., 2023). The contribution of genomics and bioinformatics towards these targets, and many of the associated technical and scientific challenges are described in Waterhouse et al. (2022), together with the possible contribution of the ELIXIR European Strategy Forum for Research Infrastructures to meet them.
Biodiversity researchers are increasingly realising the potential offered by modern technologies, particularly in genomics, to create new opportunities for developing tools and resources that will transform the field. These opportunities lie primarily in the types of scientific applications that are becoming more feasible and scalable through continued advances in genomics technologies alongside enhanced data management systems. A long-term vision sees a future where sequence-based biodiversity monitoring at scale becomes a default and provides the means for ecosystem biodiversity characterisation in space and time, complemented and enhanced by other biomonitoring technologies. In support of realising these opportunities, ongoing global and regional efforts are building capacity to generate catalogues of reference DNA barcodes (International Barcode of Life, iBOL) (Hobern, 2021) and genomes by the Earth BioGenome Project (EBP) (Lewin et al., 2018, 2022) as well as the European Reference Genome Atlas (ERGA, 2023), or both by the Biodiversity Genomics Europe (BGE, 2023; Mazzoni et al., 2023) project. Along with this increased production, concurrent development of the necessary tools and resources will greatly enhance our abilities to:
• Maintain and query increasingly comprehensive reference DNA barcode and genome catalogues, improving taxonomic coverage and differentiation (including of cryptic species), and coordinating the efforts of various initiatives under global and regional umbrellas e.g., McGee et al. (2019);
• Connect and integrate these molecular resources with other biodiversity data (traits, observations, literature, etc.) e.g., König et al. (2019), using an increasingly standardised and harmonised taxonomic framework as the common backbone;
• Use these integrated resources for applied data-driven science to understand the diversity of extant life on Earth, how that diversity functions and interacts, and how it responds to changing environmental pressures (Pereira et al., 2012);
• Implement monitoring of lesser-known or complex ecosystems, including for enhancing understanding of species interactions and dynamics, as well as for species discovery and exploration of “dark taxa” e.g., Rahman et al. (2022);
• Include assessments of within-species, population-level genetic diversity to support characterisations of their evolutionary histories and predictions of their future prospects in the face of ongoing climatic changes (Pearman et al., 2024);
• Operationalise the assessment of Essential Biodiversity Variables (EBVs) across taxa and spatiotemporal scales, focusing on species distribution and abundance (Kissling et al., 2018; Jetz et al., 2019);
• Engage with naturalists and citizen scientist groups through the use of new technologies that help build a democratised monitoring framework and improve characterisation of ecosystem biodiversity in space and time (Robinson & Peres, 2021);
• Evaluate biodiversity declines, as well as population-level adaptation and migration processes, in the context of anthropogenic activities (e.g., climate change and urbanisation consequences, (Finn et al., 2023)), and understand key aspects necessary to restore ecosystem functions (Breed et al., 2019) to help prioritise biodiversity conservation, restoration, and “rewilding” efforts (e.g., particularly relevant to at-risk biodiversity hotspots).
The field of biodiversity assessment and research, from an organisational context, is broad, complex, and distributed. There are a multitude of organisations that operate across international borders, within countries, and at a local level (see Extended Data (Waterhouse, 2023)). This landscape is further demarcated along scientific and technical lines, with organisations that focus on taxonomies, ecology, molecular sciences, and method development (necessitated by the increasingly large and complex amount of data being generated). ELIXIR, perhaps uniquely, stands as a hub for the molecular sciences and bioinformatics at an international and national level across many scientific disciplines (Waterhouse et al., 2022). Biodiversity research and infrastructures increasingly rely on molecular data (Karp et al., 1997; Porter & Hajibabaei, 2018), so ELIXIR is well placed to lead organisational alignments and collaborations: from a core set of partners across Europe mainly within the field of molecular sciences, to an expanding variety of partner organisations that focus on other biodiversity-related research and resources (see below for examples from the ecosystem of biodiversity projects, resources, and infrastructures). Importantly, this extends beyond the data themselves as FAIRification of digital research objects (Wilkinson et al., 2016), championed by ELIXIR’s Services and Platforms, is increasingly recognised as essential in biodiversity research (Wetzel et al., 2018; Lannom et al., 2020). Opportunities to help coordinate and align organisational activities in the biodiversity domain arise naturally from ELIXIR’s established European-wide “network of networks” approach, connecting to existing initiatives at both the national and international levels. With ELIXIR’s strengths in molecular sciences, a “hub and spokes” model would help augment opportunities to connect molecular-focused bioinformatics tools, protocols, and resources with the many other biodiversity-related infrastructure and stakeholder organisations. Building on these strengths in data science and a connected network across Europe, ELIXIR can contribute to coordinated efforts designed to support and grow the many existing initiatives in the domains of biomonitoring, ecosystem health, and biodiversity research.
The variety of existing biodiversity data infrastructures and resources is a testament to the long-standing recognition by multiple stakeholders of their importance, currently reflected in the growing European and global commitments to prevent further biodiversity decline and ensure the long-term health of ecosystem services. This heterogeneity, however, gives rise to many challenges, both technical in terms of data analysis (due to inadequacies of existing methodologies), data integration and data interaction, and at the level of the scientific community, which faces a heterogeneous landscape of infrastructures and resources that can be difficult to navigate (Blaxter & Floyd, 2003; Huang et al., 2012; Levin et al., 2014). The methodological and logistical challenges range from scaling up (needed to be able to process the increasing amounts of complex molecular data) to the management of these data and working on connecting them to other biodiversity research infrastructures (Waterhouse et al., 2022). The biodiversity research community needs to proactively seek common solutions that enable molecular technologies to advance biodiversity research. A key part of this is the building of distributed infrastructures for life-science data that avoid or minimise unnecessary duplication of effort to be able to advance efficiently towards common goals. To this end, informatics solutions will need to be developed to address the practicalities of common challenges, such as:
• The need to constantly incorporate knowledge-based updates and resolve conflicts to maintain standardised taxonomies that serve as a dynamic framework that facilitates interoperability across research infrastructures;
• Building data and metadata brokering services that support coordinated community engagement to ensure good data management through technical infrastructures for aiding and automating data submission;
• Developing the means, through text mining and curation, to identify and liberate in digital form invaluable historical or baseline data trapped in the literature (including those published in non-English sources), or in museum and other natural history collections;
• Improving the accessibility of research results through publications (e.g., by making published traits, tables, treatments, specimens, figures etc.), citable and reusable (e.g., through nanopublications), and including identifiers of cited elements (genes, specimens, taxonomic names, treatments);
• Improving and harmonising currently highly heterogeneous metadata collection standards to promote the adoption of community best practices that will maximise findability, accessibility, interoperability, and reusability of digital research objects (i.e., drive biodiversity research towards FAIR compliance);
• Scaling up of services for data and metadata management to keep pace with and accommodate the increases in data production (e.g., genomics) and collection (e.g., Essential Biodiversity Variables);
• Developing frameworks that deliver an increasingly integrated and interconnected landscape of biodiversity research infrastructures, utilising developments in application programming interfaces and Semantic Web services;
• Ensuring widespread access to high-performance computing (HPC) and HPC-deployable software and data-management systems, including containers and workflows, to enable decentralised efforts while promoting standardisation.
ELIXIR Communities are groups of experts across ELIXIR Nodes and beyond that represent a scientific or technological theme which drives the development of standards, services, and/or training in and across services offered by ELIXIR, thereby connecting the infrastructure services to research domains (Heil & Garrard, 2024). The ELIXIR Biodiversity Community was first launched in 2019 as a Focus Group to develop and coordinate ELIXIR Nodes’ tools, resources, and research work connected to the biodiversity domain. As part of the process of maturing from a Focus Group to a Community, members initiated activities including: (1) cataloguing ELIXIR Services that support biodiversity research; (2) developing and publishing their “Recommendations for connecting molecular sequence and biodiversity research infrastructures through ELIXIR” (Waterhouse et al., 2022); (3) coordinating ELIXIR Node participation in Horizon Europe project proposals - The Biodiversity Community Integrated Knowledge Library (BiCIKL) and Biodiversity Genomics Europe (BGE); and (4) beginning to establish connections with key external partners/projects in the biodiversity domain (such as those listed in Table 1); leading to the formal recognition in 2022 as an ELIXIR Community (Waterhouse et al., 2023).
| Project | Node/Funder | Summary details/description |
|---|---|---|
| ARISE | Netherlands | ARISE (Authoritative and Rapid Identification System for Essential biodiversity information) is a digital infrastructure with a mission to provide semi-automated identification of all multicellular species in the Netherlands (van Ommen Kloeke et al., 2022). |
| BiCIKL | E.C. (coordinated by Pensoft) | BiCIKL (Biodiversity Community Integrated Knowledge Library) will catalyse a culture change in the way biodiversity data is identified, linked, integrated and re-used across the research cycle. We will cultivate a more transparent, trustworthy and efficient research ecosystem. |
| Biodiversity Genomics Europe (BGE) | E.C. (coordinated by Naturalis Biodiversity) | By bringing together Europe’s key practitioners in two fundamental DNA-based technologies - DNA barcoding and genome sequencing - the BGE consortium aims to streamline the rollout of these methods across Europe. |
| Biodiversity Digital Twin (BioDT) | E.C. (coordinated by CSC – IT CENTER FOR SCIENCE LTD.) | The Biodiversity Digital Twin prototype will provide advanced models for simulation and prediction capabilities, through practical use cases addressing critical issues related to global biodiversity dynamics. |
| Curated collections of DNA barcode marker | Italy | A reference collection of COXI mitochondrial DNA genes based on the integration of sequence and taxonomy data of BOLD and ENA (Balech et al., 2022). |
| e-BioDiv | Switzerland | Open Biodiversity FAIR-ification Services for Biospecimens stored in Swiss Natural History Museums |
| Earlham Institute Barcoding the Broads | UK | A Wellcome-funded programme of public engagement events and activities to explore biodiversity on the Norfolk Broads, led by the Earlham Institute as part of the work on the Darwin Tree of Life project. |
| ELIXIR Norway | Norway | Dedicated national ELIXIR Node funding (2022-2026) includes a focus on biodiversity and connections to other biodiversity infrastructures and projects in Norway (e.g., the Earth BioGenome Project Norway: EBP-Nor). |
| Establishment of an ELIXIR Contextual Data Clearinghouse | ELIXIR (Implementation study) | The objective is to develop and deploy an “ELIXIR Contextual Data Clearinghouse” for extending, correcting and improving publicly available annotations on records in sample and sequencing data resources. |
| Molecular Biodiversity Greece Community (MBGC) | Greece | Greece is a biodiversity hotspot and to this end, a network of networks covering different disciplines of molecular biodiversity research has been developed. MBGC aims to channel the flow of information amongst researchers, institutions, policy makers, stakeholders and local communities, remaining aligned to all relevant initiatives and infrastructures, at the national, European, and global level. |
| NFDI4Biodiversity | Germany | Network of diverse biodiversity data (not only molecular). Data are provided by research organisations and projects (e.g., GBOL), public authorities, professional societies and citizen initiatives. Data Management oriented. The production of the data itself is done through use cases. |
| Phylogenetic methodology | Ireland | A range of analytical tools is being developed to augment the bioinformatics tool kit for comparative genome analysis. |
| Pole National de Données de Biodiversité | France | National centre of data on biodiversity: the data are provided by the same diversity of channels as in Germany and the role of PNDB is to support FAIR data management. |
Operationally, monthly online meetings coordinated by the Community co-leads with support from the ELIXIR Hub serve as the primary forum for interactions, complemented by discussions and notifications on the ELIXIR Slack Workspace’s Biodiversity Community channel. These include sharing information on members’ participation in ongoing or planned biodiversity-related projects and initiatives, including the Community-led Implementation Study “Biodiversity Networks for ELIXIR”. The online meetings also feature presentations on tools and services developed by ELIXIR Nodes as well as hosting invited speakers representing key external partners/projects. The Community’s Implementation Study encompasses four key areas of work to drive Community activities: (1) to survey and catalogue Research Data Management (RDM) elements relevant to the biodiversity domain, with a focus on molecular data; (2) to catalogue, review, and categorise tools, services, and analytical workflows currently in use by ELIXIR Nodes and the wider community, that process and analyse biodiversity-related data; (3) to describe the landscape of stakeholders ELIXIR is working with or needs to better engage with to establish a “network of networks” for biodiversity research and services; and (4) to leverage the strengths of ELIXIR’s training experience to help support the growth of the Biodiversity Community through network-driven sharing of training experiences and knowledge transfer and materials. Together, these actions are serving to enhance ELIXIR’s network of networks in helping to deliver connected data to advance biodiversity research.
Tackling the biodiversity crisis at a general level is not going to be resolved through a single action, but instead requires a complex set of interacting actions that are co-dependent but usually funded separately. ELIXIR can assume a key leading role in a subset of those actions, focused on data management and the molecular sciences, where even at the level of ELIXIR, there are a multitude of funded projects at a transnational, national, and local level. In terms of informatics solutions connected to such projects, the ELIXIR Biodiversity Community is guided by themes emerging from surveying approaches by which molecular technologies are helping to inform understanding of biodiversity (Waterhouse et al., 2022): biodiversity-related and informatics infrastructures need to develop close and strategic collaborations; work on taxonomy needs to be better aligned and standardised across different infrastructures and fields of study; metadata urgently needs harmonisation and common approaches to research data management must be widely adopted; current data science solutions need to be scaled up to address the rapidly accumulating amounts of molecular data; bioinformatics support for biodiversity research needs to be made widely available and properly maintained; user training on biodiversity research tools, services, and infrastructures needs to be prioritised; and community initiatives need to be collaborative, proactive, and solution-driven. These themes come together in a complex network of interacting projects that have distinct but related aims, usually focused on establishing communities and connections and/or building new technical solutions to help with data access, storage, or analysis. ELIXIR can serve a critical function here, as a fundamental aspect of its mission is to make connections and coordinate across complex activities. Table 1 lists a subset of ongoing projects across Europe and within ELIXIR member states that illustrate the breadth of activities underway.
ELIXIR as a Research Infrastructure is structured around (technological) Platforms as well as (user) Communities. Both of these interact on an ongoing basis, mutually supporting each other’s efforts. The ELIXIR Biodiversity Community is already collaborating with some of these and aims to broaden interactions to fully leverage the available potential and resources. Some examples of current and future interactions with ELIXIR Platforms (Tools, Compute, Data, Training, and Interoperability) are:
• The Tools Platform provides services for finding software tools and web portals (Bio.tools (Ison et al., 2019), including the https://biodiversity.bio.tools subdomain to be populated by the ELIXIR Biodiversity Community), software containers (BioContainers (da Veiga Leprevost et al., 2017)), and workflows (WorkflowHub (Goble et al., 2021)); for assessing tools (OpenEBench (Capella-Gutierrez et al., 2017)); and the best practices in providing research software (Jiménez et al., 2017)). EDAM ontology enables annotation and search of tools and other research objects by application domain, task, or data (Black et al., 2022); and an extended coverage of biodiversity research concepts could be achieved via engagement with the Biodiversity Community.
• Specifically for the Compute Platform: User accessible compute, potentially controlled user access via Authentication and Authorisation Infrastructure (AAI).
• Community data-management support, and integration with ELIXIR Core and Deposition Data resources. The European Nucleotide Archive (ENA) is a critical data deposition resource for biodiversity genomics data. A concrete example of metadata management workflow is that developed between biodiversity scientists, the Data Platform, and the Biodiversity Community Integrated Knowledge Library (BiCIKL) project (Penev et al., 2021, 2022): a metadata management workflow employs the PlutoF tool for biodiversity data and metadata management (Abarenkov et al., 2010), and the ELIXIR Data Platform services.
• Networks of tool/infrastructure users and developers to augment the Training Platform offerings (e.g., with specific courses covering aspects such as: genome annotation, meta-data brokering, etc.) and more complete learning paths, covering entire workflows (e.g., from sequencing to annotation, possibly covered via Galaxy).
• A growing necessity in the biodiversity field towards connected data, as championed by the Interoperability Platform, concretely touching on resources like: RO-Crate and link to specimens, RDMkit, FAIRsharing, Bioschemas and the FAIRcookbook. The ELIXIR Biodiversity Community aims to bring together researchers producing the data, in all their varied forms, with informaticians developing interoperability solutions, to help overcome the challenges of data heterogeneity in the field.
Regarding links between the ELIXIR Biodiversity Community and other ELIXIR Communities, these are already foreseen, and a number of synergies have been clearly identified. Some examples can be found in Table 2.
| Community | Shared activities |
|---|---|
| Food & Nutrition | Conceptualisation and implementation of interoperability data models able to integrate, standardise and harmonise data from different disciplines: metagenomics, metabolomics and transcriptomics. |
| Galaxy | Thousands of tools, including hundreds for biodiversity and microbial/microbiome analysis, are ready to be used on publicly-accessible HPC resources, together with workflows for data processing, which can be versioned, annotated, and shared for reuse. The European Galaxy server (https://usegalaxy.eu) offers access to 2700+ tools and workflows. Galaxy-Ecology is its subdomain piloted by the French ELIXIR Node. A training material repository (https://training.galaxyproject.org) is open for everyone to use and contribute to, providing slides, hands-on tutorials, and other material on using Galaxy to analyse data, with 260+ tutorials in 20+ topics including ecology, microbiome, and climate. Integration of PlutoF and other biodiversity tools into Galaxy could be carried out together with the Biodiversity Community in the near future. |
| Microbiome | Meta-genomic workflows and data archiving. Marine sample metadata annotation guidelines. |
| Plant Science | Taxonomy framework; coherent/consistent metadata standards for samples (see also interoperability PF (platform), MIAPPE (Minimum Information About Plant Phenotyping Experiments)). Alignment between the MIAPPE standard and exchange formats and the relevant TDWG (Biodiversity Information Standards) standards and exchange formats. Integration and linking different plant data types. |
ELIXIR entered the European Strategy Forum for Research Infrastructure’s (ESFRI) first roadmap in 2006 and reached its Landmark status in 2016 (ELIXIR, 2021). As a distributed research infrastructure, ELIXIR coordinates, integrates, and sustains bioinformatics resources across European countries and helps address the Grand Challenges across life sciences, from marine research, via plants and agriculture, to health research, medical sciences, and biodiversity informatics. ELIXIR provides services in seven scientific domains including “Evolution and phylogeny” and “Genes and genomes” (https://elixir-europe.org/services) that link the activities of the ELIXIR community to the wider landscape of life-science research infrastructures (RIs) and international projects. As RIs mature and FAIRness has become the standard to achieve interoperability between RIs, it is opportune to outline the global network of interrelated projects and infrastructures, in which ELIXIR operates to maximise synergy and to avoid redundancy.
The relationships between different aspects of biodiversity data are well captured by the biodiversity knowledge graph of Roderic Page (Figure 1). The key activities of ELIXIR are captured by the molecular domain; the biodiversity knowledge graph clearly indicates how molecular data are related to the wider spectrum of biodiversity data that are targeted by other RIs and projects. The ELIXIR Biodiversity Community benefits from connections to RIs and projects in the biodiversity domain, an overview of which can build on the landscape analyses of the ESFRI roadmaps of ESFRI 2018 (ESFRI, 2018) and 2021 (ESFRI, 2021), the partners of the Alliance for Biodiversity Knowledge, and the research infrastructure contact zones analysis between 10 biodiversity infrastructures, including ELIXIR (Smith et al., 2022). Additional to the data types considered by Page (Figure 1), the contact zones analysis considers ‘observations’ and ‘collections’, or groups of specimens, as elements of the biodiversity data domain. This recognition of the variety of types of biodiversity data and the importance of integration has been key to the establishment of many RIs and research projects, for example: the Alliance for Biodiversity Knowledge; Biodiversity Genomics Europe; Biodiversity Heritage Library; Biodiversity Community Integrated Knowledge Library; iBOL BIOSCAN; Biodiversity Literature Repository; Catalogue of Life; Data Observation Network for Earth; Distributed System of Scientific Collections; Earth BioGenome Project; European Marine Biological Resource Centre; Environmental Research Infrastructures; Encyclopedia of Life; European Open Science Cloud; European Reference Genome Atlas; Europa Biodiversity Observation Network; Global Biodiversity Information Facility; Global Earth Observation System of Systems; Global Soil Biodiversity Initiative; International Barcode of Life; iNaturalist; LifeWatch ERIC; Long-Term Ecosystem Research in Europe; Microbial Resource Research Infrastructure; National Ecological Observatory Network; Open Traits Network; Plazi; Pôle national de données de biodiversité; Swiss Institute for bioinformatics Literature Services; Soil Biodiversity Observation Network; TreatmentBank; World Register of Marine Species.
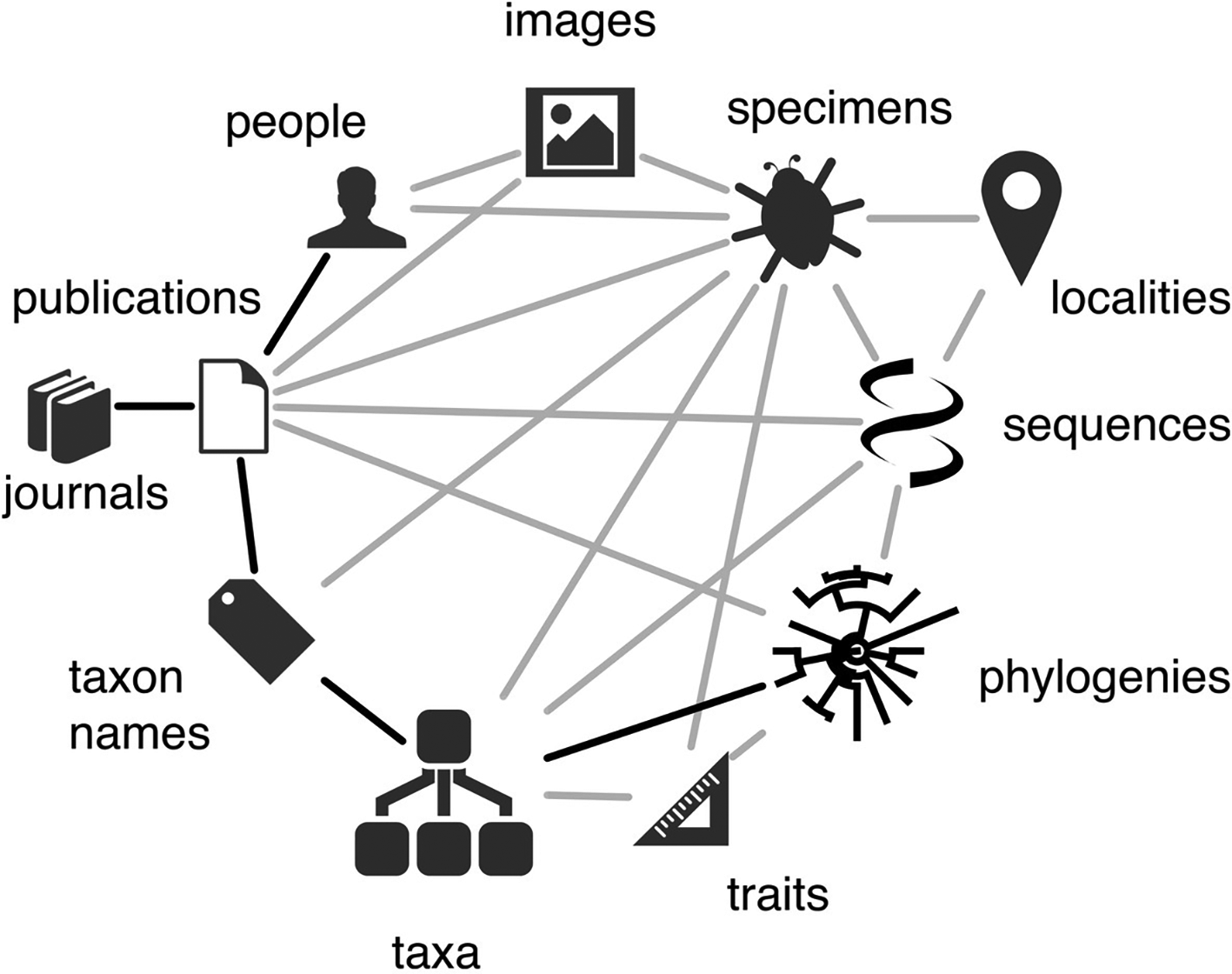
Genomics data comprise one facet of the biodiversity knowledge graph, where questions and approaches in biodiversity research traverse the paths in this graph, and where all parts of the graph are constantly ‘evolving’ and growing. Wikimedia Commons CC-BY-4.0.
In addition to the above examples of biodiversity projects and infrastructures that make up the global network of ongoing efforts in the domain, and in the context of ELIXIR’s participation and contribution to the network, the following two examples highlight ongoing activities in the field of biodiversity and in the context of the European research sphere.
Several ELIXIR Nodes are involved in European projects with a focus on biodiversity. The BiCIKL project is building the Biodiversity Knowledge Hub (BKH) - a single knowledge portal to interlinked machine-readable FAIR data - using unique stable identifiers on specimens, sequences, taxonomy and publications (Penev et al., 2021, 2022). A set of core global biodiversity databases (GBIF, ENA, PlutoF, Plazi, DiSSCo, OpenBioDiv, ToL, etc.) are contributing with the aim to develop services to augment the interlinking of biodiversity contents, starting with biotic interactions. The project is also financing competitive implementation studies to develop transnational resources.
The European Open Science Cloud initiative (2023) intends to offer a federated and open multi-disciplinary environment where tools, data and services can be published, sought, and re-used. Via enabling seamless access and FAIR management EOSC aims to develop a Web of FAIR Data and services for science, innovation and education in Europe through which value-added services can be offered. The EOSC-Life initiative connects 13 life science ‘ESFRI’ research infrastructures to create an open, digital and collaborative space for biological and medical research. Among the EOSC-Life “FAIR” published data and catalogued services (by participating RIs), ones related to biodiversity are included. The workflow for marine Genomic Observatories data analysis is such an example (EBI, 2021).
Considering the context discussed above in terms of the complex landscape of ongoing initiatives working to meet the most pressing needs supporting biodiversity research and services, the ELIXIR Biodiversity Community aims to contribute towards the global aim of tackling the biodiversity crisis by helping to make possible a future where:
• Large-scale sustainable data production services are meeting the routine needs of hundreds of laboratories and thousands of citizen scientists for sequence-based biodiversity research and biomonitoring;
• ELIXIR is part of a network of well-connected, stable, and long-term infrastructures that is supporting a growing portfolio of stakeholders in biodiversity research by improving their access to, and integration of well-curated, high-quality, richly annotated, and connected molecular data.
• State-of-the-art computational tools and services are available for large-scale projects related to biodiversity, including data standardisation initiatives, allowing for the enhanced exploitation of the collected and connected data within the biodiversity knowledge graph.
To define a roadmap for the ELIXIR Biodiversity Community to help drive progress towards advances in these three overarching priority areas – data production, data integration, and data exploitation – a set of five guiding goals has been established:
1. To enhance ELIXIR’s network of networks in helping to deliver connected data for biodiversity research by: exposing and augmenting relevant ELIXIR services and infrastructures contributing to efforts to catalogue, protect, and restore biodiversity and ecosystem services; supporting ELIXIR Nodes in expanding their activities related to biodiversity data and research and relevant for the new programme priority areas; and communicating ELIXIR activities and services relevant to the biodiversity domain to broader audiences including scientists, policy makers, and the general public.
2. To support the development of standards and promote best practices in biodiversity research by: supporting and promoting the development and use of global standards, formats, guidelines, and ontologies across the domain; supporting the development of best practices in Research Data Management for biodiversity research, with a focus on molecular data but not excluding other areas; and collecting, exposing, and maintaining, a dedicated RDMkit domain for biodiversity collating relevant documentation and tools that support good practices in research data management.
3. To promote tools and workflows that facilitate reliable and reproducible biodiversity data analyses by: identifying, curating, and promoting high-quality biodiversity-related analysis tools and services to the wider community of users; connecting developers with data-generation initiatives and data users to fuel synergies that deliver tools and workflows serving their needs; and maintaining curated catalogues of FAIR biodiversity-focused tools and workflows in Bio.tools and WorkflowHub.
4. To enhance biodiversity database/infrastructure usability and interoperability by: identifying and acting on opportunities to develop technical solutions that improve connectivity amongst heterogenous biodiversity data infrastructures and resources; fostering collaborative exchanges between users and providers of biodiversity-related databases/infrastructures to improve usability and functionality; and promoting the usage of persistent identifiers across the domain, including taxonomies as the backbone of biodiversity knowledge.
5. To foster knowledge transfer in biodiversity data management and analysis by: supporting community-driven skills sharing focused on understanding how to benefit from the use of available standards and best practices; connecting developers of tools/workflows/databases with user communities through training that responds to changing technologies and associated services; and exposing collections of training materials, for example through TeSS, the Galaxy Training Network, and RDMkit.
To complement these guiding goals, the ELIXIR Biodiversity Community proposes a roadmap towards achieving our long-term objectives. Table 3 shows five long-term objectives for the ELIXIR Biodiversity Community to address, centred on engaging with stakeholders, aligning infrastructures, contributing to policy, supporting production, and enhancing ELIXIR’s portfolio in the biodiversity domain. The current focus is on the informatics, databases, and tools more than on the biological questions, so as the Community grows, it will be important to widen the diversity of its membership to ensure that the technical developments will serve the needs of biodiversity researchers. Beyond the ELIXIR Biodiversity Community itself, it is also vital to engage with other communities in the domain, including with stakeholders such as practitioners and citizen science initiatives in order to contribute towards bridging the gaps between research and implementation (Dubois et al., 2020; Fraisl et al., 2022).
Figshare: Extended Data 1: Biodiversity RIs & Projects. https://doi.org/10.6084/m9.figshare.22723432 (Waterhouse, 2023).
This project contains the following extended data:
- Extended_Data_1_Biodiversity_RIs_Projects.xlsx (A non-exhaustive list of biodiversity research infrastructures, collected as part of the development of the ELIXIR Biodiversity Community white paper 2022-2023.)
Data are available under the terms of the Creative Commons Zero “No rights reserved” data waiver (CC0 1.0 Public domain dedication).
The authors thank all members of the ELIXIR Biodiversity Community, the ELIXIR Hub, and the ELIXIR Heads of Nodes, for useful feedback during the development of this Community White Paper, and the reviewers for their invaluable input.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: biodiversity informatics, open science, data standards
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: macroecology, biodiversity data, ecoinformatics, marine ecology
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Biodiversity Conservation, Landscape Ecology, Biodiversity status assessment, Climate change vulnerability and adaptation
Is the topic of the opinion article discussed accurately in the context of the current literature?
Yes
Are all factual statements correct and adequately supported by citations?
No
Are arguments sufficiently supported by evidence from the published literature?
Partly
Are the conclusions drawn balanced and justified on the basis of the presented arguments?
Yes
References
1. Pereira H, Navarro L, Martins I: Global Biodiversity Change: The Bad, the Good, and the Unknown. Annual Review of Environment and Resources. 2012; 37 (1): 25-50 Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: macroecology, biodiversity data, ecoinformatics, marine ecology
Is the topic of the opinion article discussed accurately in the context of the current literature?
Partly
Are all factual statements correct and adequately supported by citations?
Partly
Are arguments sufficiently supported by evidence from the published literature?
Partly
Are the conclusions drawn balanced and justified on the basis of the presented arguments?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: biodiversity informatics, open science, data standards
Is the topic of the opinion article discussed accurately in the context of the current literature?
Yes
Are all factual statements correct and adequately supported by citations?
Yes
Are arguments sufficiently supported by evidence from the published literature?
Yes
Are the conclusions drawn balanced and justified on the basis of the presented arguments?
Partly
References
1. Harrow J, Hancock J, ELIXIR-EXCELERATE Community, Blomberg N: ELIXIR-EXCELERATE: establishing Europe's data infrastructure for the life science research of the future.EMBO J. 2021; 40 (6): e107409 PubMed Abstract | Publisher Full TextCompeting Interests: No competing interests were disclosed.
Reviewer Expertise: Biodiversity Conservation, Landscape Ecology, Biodiversity status assessment, Climate change vulnerability and adaptation
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 22 May 24 |
read | read | read |
|
Version 1 15 May 23 |
read | read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)