Keywords
Haemagogus equinus, mitogenome, Jamaica, arboviruses, genome skimming
This article is included in the Genomics and Genetics gateway.
In the Americas, the expansion in incidence of arboviral infections including Mayaro virus (MAYV) has drawn attention to the resurgence of viruses associated with understudied arthropods. Mosquitoes belonging to the genus Haemagogus are generally geographically restricted to the forests of Central and South America and the Caribbean and are the known sylvan vectors for yellow fever virus and emerging MAYV. With an established population in Jamaica, Haemagogus equinus has been reported to be well-adapted to oviposition in artificial containers close to human populations. Its role in arboviral transmission however is not fully understood. Given the dearth of genetic information and the difficulty in morphologically identifying cryptic features in species belonging to this genus, we report the first mitochondrial genome of Hg. equinus. Using a genome skimming approach, two Hg. equinus mosquito specimens were sequenced using the Illumina Novaseq 6000 platform. A representative mitogenome of 16,471 bp, 80.7% AT and 37 genes was assembled using NOVOplasty. Phylogenetic analysis placed Hg. equinus in the Albomaculatus section of the Haemagogus subgenus supporting previously described taxonomic studies.
Haemagogus equinus, mitogenome, Jamaica, arboviruses, genome skimming
The revised version of this article now includes additional details regarding the molecular analysis which was conducted. Furthermore, a higher resolution version of figure 1 has been provided and figure 2 has been revised with the addition of Ae. aegypti as an outgroup in the phylogenetic tree, and the Haemagogus and Conospotegus subgenera are now both clearly displayed in the figure. Minor revisions have also been made to the figure 2 legend to provide further clarity regarding the use of Psorophora and Aedes as outgroups for the phylogenetic tree. Information pertaining to the similarity between the two sequenced specimens has also been added to the results section.
See the authors' detailed response to the review by Craig Wilding
See the authors' detailed response to the review by Din Syafruddin and Puji Budi Setia Asih
Mosquitoes of the genus Haemagogus (Hg.) Williston, 1896 are endemic to tropical rainforests, open deciduous forests and mangroves in Latin America and the Caribbean.1 The genus is divided into two subgenera (Haemagogus and Conopostegus), with the Haemagogus subgenus split into three sections (Albomaculatus, Splendens, Tropicalis).1 Being the primary sylvatic vectors for yellow fever virus (YFV)1 and the emerging arboviral threat Mayaro virus (MAYV),2 species in this genus of mosquitoes are of significant medical importance in the region. Despite this, many species remain understudied and their role in disease transmission has not been clearly defined. In Jamaica, Haemagogus equinus Theobald, 1903 is presently the only known species on the island. Established populations of Hg. equinus have also been identified in southern Texas in the United States of America, Mexico, Belize, Guatemala, El Salvador, Honduras, Nicaragua, Costa Rica, Panama, Colombia, Guyana, Venezuela and Trinidad and Tobago.1
While the vectorial capacity of Hg. equinus has not been fully elucidated, laboratory and transovarial transmission of YFV3–6 along with natural infections among wild mosquitoes have been reported.7,8 In spite of this, its importance in arboviral transmission in some localities remains uncertain.9 Although Hg. equinus primarily oviposits and larvae develop in tree holes and bamboo internodes,1 the species is very adaptable and can also utilize rock holes,10 domestic containers and used tires.11,12 With multiple reports of MAYV in neighbouring Haiti,13–15 there is need for a greater understanding of the role of this sylvatic vector in arbovirus transmission. To facilitate this, accurate identification of all life stages of field collected specimens is crucial. However, impediments such as a lack of taxonomic expertise coupled with poor quality samples and a paucity of reference molecular data continue to hamper these efforts in Jamaica.
Few studies have employed genomic sequencing to investigate the molecular taxonomy and phylogenetic relationships of Hg. equinus mosquitoes. Currently, only partial sequences of cytochrome oxidase I and other mitochondrial or nuclear genes are available from NCBI’s GenBank or BOLD systems databases.16–21 The complete mitochondrial genome (mitogenome) is increasingly being used for evolutionary and phylogeny studies due to its maternal inheritance, simple genomic organization, relative abundance in tissues and absence of recombination.22 In most metazoans the mitogenome is highly conserved and comprises of 13 protein coding genes, 22 transfer RNAs and two ribosomal RNAs in addition to a large single non-coding region important for replication and transcription.23 At present, mitogenomes of only five species from the genus Haemagogus are available.24,25 Considering the significance of mitogenomes when conducting phylogenetic and taxonomic studies, we describe for the first time the characterization of the mitogenome of Hg. equinus using a genome skimming approach and the phylogenetic comparison with other Haemagogus taxa.
The Hg. equinus samples (n = 2) sequenced were collected in Mona, Jamaica (18.0061364°N, -76.7515125°W) in May and December 2023 in an area characterized by the predominant growth of banana plants and adjacent to a forested area. Briefly, BG sentinel traps (BioGents, Regensburg, Germany) baited with two pounds of dry ice (without lure) were placed overnight from 1400 to 1000 hr and collected mosquito specimens were sorted and morphologically identified using taxonomic keys.26 Single specimens were stored in tubes containing silica and shipped to the Johns Hopkins Bloomberg School of Public Health for molecular analysis as described in detail in.27 Briefly, each mosquito specimen was homogenized in PK buffer (Applied Biosystems, Waltham, USA) containing proteinase K (Applied Biosystems, Waltham, USA) and incubated at 56°C. DNA was then extracted using the Qiagen DNeasy Blood and Tissue Kit (Hilden, Germany) and quantified using the Qubit dsDNA assay kit (ThermoFisher, Waltham, USA) prior to library construction and Illumina sequencing which was conducted at SeqCenter (Pittsburgh, USA).
The mitogenome was assembled as described for African anophelines using NOVOPlasty (RRID:SCR_017335) version 4.3.127,28 and automatic annotations performed with MITOS on the galaxy platform under the invertebrate genetic code.29,30 Geneious Prime (RRID:SCR_010519) version 2023.2.1 (Biomatters, Auckland, Australia) was utilized for manual adjustments of start and stop codons to match reference Haemagogus mitogenomes in the GenBank repository. Figure 1 illustrates a representative map of the mitochondrial sequences and annotations submitted to GenBank.
Nine mitogenomes (two sequenced from this study and seven available from GenBank) were used for phylogenetic analysis. The 13 protein coding genes of these mitogenomes were extracted with Geneious Prime and aligned by MAFFT version 7.490 into a single matrix. jModelTest version 2.1.10 identified the best fit substitution model and phylogenetic analyses were performed using maximum likelihood in MEGA (RRID_SCR_023017) version 11 with 1000 bootstrap replicates.
Sequencing of the two Hg. equinus specimens resulted in a mean of 26,728,491 million reads and of these approximately 69,261 reads were utilized for assembling each mitogenome. The two specimens shared 98.84% similarity. Two ribosomal RNAs, 22 transfer RNAs and 13 protein coding genes were detected in the two Hg. equinus mitochondrial genomes (GenBank accession numbers PQ_189398, PQ_189399). The cytochrome c oxidase I (COI) segment covering 1691-3221 bp was 99.59 % comparable to a COI sequence for Haemagogus spegazzinii Brèthes, 1912 (YP_010155459) retrieved from GenBank. Similar to the six Haemagogus mitochondrial genomes available in the GenBank database, the representative mitogenome from our sequencing efforts (PQ_189398) has an A+T proportion of 80.7% and length of 16,471 bp. The Maximum Likelihood phylogenetic tree places it in the Albomaculatus section, separated from other Haemagogus mitogenomes ( Figure 2).
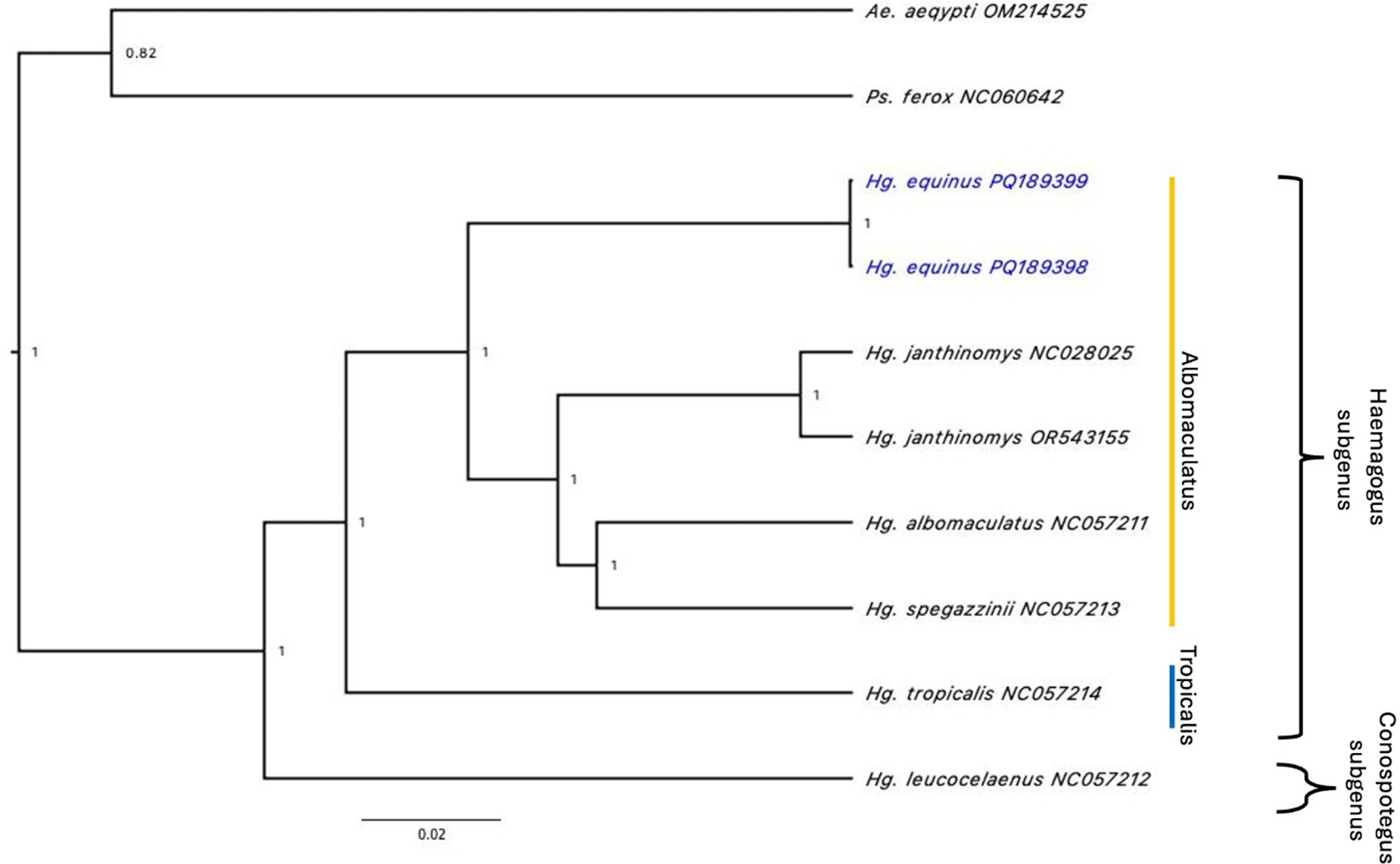
These findings provide the basis for the development of more accurate molecular tools which can be used for identification of Hg. equinus mosquitoes. Furthermore, by populating molecular repositories with genomic data from Jamaican mosquitoes, their phylogenetic and evolutionary status will be better understood.
GenBank: Haemagogus equinus mitochondrion, complete genome. Accession numbers PQ189398, PQ189399; https://identifiers.org/ncbi/insdc:PQ189398, https://identifiers.org/ncbi/insdc:PQ189399.31
Bio Project. Complete mitochondrial genome of Haemagogus equinus, Accession number PRJNA1172362; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1172362.32
SRA. Illumina seq of Haemagogus equinus. Accession numbers SRR31002245, SRR31002246; https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA1172362.33
Biosample: Haemagogus equinus isolates HGJ1, HGJ3. SAMN44269491, SAMN44269492; https://trace.ncbi.nlm.nih.gov/Traces/study/?acc=PRJNA1172362.34
Simmoy A.A. Noble is a Global Infectious Diseases Scholar and received mentored research training in the development of this manuscript.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Comparative genomics, chromosome evolution
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
No
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Evolutionary genetics
Are the rationale for sequencing the genome and the species significance clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of the sequencing and extraction, software used, and materials provided to allow replication by others?
Yes
Are the datasets clearly presented in a usable and accessible format, and the assembly and annotation available in an appropriate subject-specific repository?
Yes
Competing Interests: No competing interests were disclosed.
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | |||
|---|---|---|---|
| 1 | 2 | 3 | |
|
Version 2 (revision) 02 Jul 25 |
read | ||
|
Version 1 09 Dec 24 |
read | read | |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)