Keywords
Word embedding, drug repurposing, SARS-CoV-2, COVID-19
This article is included in the Emerging Diseases and Outbreaks gateway.
This article is included in the Bioinformatics gateway.
This article is included in the Coronavirus (COVID-19) collection.
Word embedding, drug repurposing, SARS-CoV-2, COVID-19
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and associated coronavirus disease 2019 (COVID-19) were first identified in December of 2019 and have since spread to become a global pandemic1. This rapid spread of illness and death demands a rapid response in treatment development. De novo drug development, however, is slow, expensive, and suffers from low probability of success2. In contrast, drug repurposing, identifying new indications for existing drugs, offers the advantages of reduced time and risk to finding treatments. We thus propose that drug repurposing is the most promising approach to treatment development for this pandemic.
There are several strategies we could employ for drug repurposing. Certainly, getting access to the rapidly growing electronic health record (EHR) histories of those afflicted by COVID-19 could be enlightening. We could, for example, track patient recovery times and look for common prescription histories in those who recover sooner. Gaining access to sufficient EHR data would likely prove challenging though due to privacy concerns and limited data at individual institutions, not to mention the added administrative burden that might entail for an already strained health system. Given the similarity of SARS-CoV-2 to its predecessor SARS-CoV3, we propose leveraging what we have learned about SARS in the intervening years. Specifically, we propose mining a word embedding built on biomedical literature published through early 2019 for candidate FDA approved drugs to treat SARS. Our results show that our proposed approach identifies several promising candidate drugs that have already been suggested or are already in clinical trials for COVID-19. We thus propose other candidate drugs identified by our method as potential leads for further investigation via in vitro and in vivo experimentation.
In the following sections, we describe our word embedding source, our source and processing method for FDA approved drug names, and our approach to mining the word embedding for drugs to treat SARS. We then present our results and a discussion including manual evaluation of the top candidate drugs proposed by our method, followed by a conclusion and suggestions for future work.
In order to perform our word embedding mining for COVID-19 drug repurposing, we first need a word embedding. Furthermore, we need drug names to look for within the embedding. Here we briefly describe our sources for both the word embedding and drug names, we describe the data processing we perform on these sources, and we describe our methods for analysis. Code and data used for all of this analysis can be found at https://github.com/finnkuusisto/covid19_word_embedding4.
Rather than spend the time building our own word embedding on biomedical text, we instead searched the literature where there are several prebuilt biomedical word embeddings available. For this work, we chose the BioWordVec5 prebuilt embedding, specifically the intrinsic model. We chose BioWordVec because it is the most recent available biomedical word embedding and it has performed well on several benchmark tasks.
In order to find a vector representation for COVID-19 treatments, we use a simple analogy approach. The original Word2vec publication demonstrated that the structure of a word embedding space could carry semantic meaning by showing that vector(“King”) - vector(“Man”) + vector(“Woman”) resulted in a vector closest to the word vector for Queen6. Effectively, this vector math asks the analogy King is to Man as what is to Woman? We use the same approach here, but instead use common drug-disease pairs as the seed analogy and SARS as the query disease. For example, one analogy we use is: vector(“Metformin”) - vector(“Diabetes”) + vector(“SARS”). Effectively, we get the word vector analogy of Metformin is to Diabetes as what is to SARS? Note that the BioWordVec embedding we are using was published before SARS-CoV-2 was discovered and thus contains no reference to SARS-CoV-2 or COVID-19 in the vocabulary. Given, that SARS-CoV-2 is a strain of SARS-CoV7, we use SARS as an approximation. To get a sense of analogy consistency, we use three separate drug-disease pairs as our seed treatment analogies: metformin and diabetes, benazepril and hypertension, and albuterol and asthma.
Given the urgency of the situation, we consider drug repurposing the most appropriate approach to finding treatments for COVID-19. We thus chose to tailor our treatment mining toward finding FDA approved drugs, allowing for the potential of off-label prescription in the short term. To get a list of approved drugs for our embedding analysis, we downloaded the FDA’s approved drug database8, extracted the drug names, and processed them for use in the word embedding.
To extract raw drug names from the FDA database, we first pulled all entries from the DrugName and Active-Ingredient fields of the Products table. We next manually inspected all raw entries that ended with parentheticals (e.g. “prempro (premarin;cycrin)”) to identify entries that contain aliases or combinations versus those that contain tokens related to branding or packaging (e.g. “rogaine (for men)”). From these parentheticals, we manually collected additional drug names and then removed all parentheticals from the drug entries. These manually collected additional names included Ampicillin, Cycrin, Hydrocortisone, Premarin, Sulfabenzamide, Sulfacetamide, Sulfathiazole, Sulfadiazine, Sulfamerazine, and Sulfamethazine. We then split all of the entries by the semicolon character to separate drug names and ingredients entered as lists. Finally, we manually added back in those drugs and ingredients that were manually extracted from the deleted parentheticals. This gave us a list of 8,561 candidate approved drug names.
We next converted our candidate drug names into word vectors to enable ranking by their similarity with our treatment analogy vector. Here we simply split each candidate drug by white space and averaged the individual token vectors to get a final vector for the drug overall. When a token was not present in the embedding vocabulary, we simply dropped that token from the average and from the initial drug name. We used this approach rather than dropping a drug entirely to allow greater flexibility, for example if the embedding vocabulary is missing an ingredient from a combination drug. Finally, we removed duplicate drug names with the same tokens to account for exact duplicates and those with combinations stated in multiple orders. As a result, we successfully derived 5,833 distinct drug vectors from our initial 8,561 candidate drugs. We then sort these drug vectors by cosine similarity with our treatment analogy vectors and evaluate the closest hits.
As a preliminary validation that our approach can work to find useful drugs for diseases from treatment analogy vectors, we first considered major diseases and disease families with well-known treatments. Specifically, we used our treatment analogy vector approach to rank drugs for the query diseases Alzheimer’s, allergies, and cancer (see Table 1, Table 2, and Table 3). Note that we still used the same seed drug-disease pairs here (metformin-diabetes, benazeprilhypertension, and albuterol-asthma) but searched for analogous treatments for Alzheimer’s, allergies, and cancer instead of SARS. For example, one analogy we used for initial validation is: vector(“Metformin”) - vector(“Diabetes”) + vector(“Alzheimer’s”). For this preliminary validation, we wanted to find drugs whose main indication is to treat the query disease in the top candidates. We chose these query diseases because they are fairly broad and have minimal treatment overlap with the seed drug-disease pairs that we used for the analogy. After initial validation of our method, we manually reviewed the top 50 drug candidates for SARS using the same method (see Table 4, Table 5, and Table 6).
Drugs with a primary indication for Alzheimer’s are highlighted in gray.
Drugs with a primary indication for allergies are highlighted in gray.
Drugs with a primary indication for cancer are highlighted in gray.
Hits containing drugs suggested or under investigation for COVID-19 are highlighted in gray.
| Metformin-Diabetes as ?-SARS |
|---|
| gilteritinib fumarate peramivir |
| zanamivir9 |
| erdafitinib |
| atovaquone and proguanil hydrochloride10 rimantadine hydrochloride11,12 |
| delavirdine mesylate |
| atazanavir sulfate and ritonavir13 |
| cobimetinib fumarate |
| niclosamide14 lopinavir and ritonavir13 temsirolimus15 |
| rilpivirine hydrochloride alectinib hnydrochloride lefamulin acetate |
| perphenazine and amitriptyline hydrochloride16 |
| alogliptin and metformin hydrochloride |
| tamiflu17 selinexor18 |
| amprenavir |
| ibuprofen and diphenhydramine citrate19 |
| olanzapine and fluoxetine hydrochloride |
| probenecid and colchicine20 |
| erlotinib hydrochloride |
| bicalutamide21 |
| alomide |
| amantadine hydrochloride11,12 azelastine hydrochloride and fluticasone propionate22 |
| revefenacin imipramine pamoate doravirine rosiglitazone maleate and metformin hydrochloride nefazodone hydrochloride |
| mefloquine hydrochloride23,24 |
| abacavir sulfate and lamivudine carisoprodol compound triprolidine and pseudoephedrine hydrochlorides codeine soma compound codeine |
| chloroquine hydrochloride25 saquinavir mesylate26 linagliptin and metformin hydrochloride27 |
| nilutamide |
| donepezil hydrochloride and memantine hydrochloride11,12 nelfinavir mesylate28 |
| ceritinib |
| virazole29 |
| vorinostat triprolidine and pseudoephedrine hydrochlorides fulvestrant gefitinib |
Hits containing drugs suggested or under investigation for COVID-19 are highlighted in gray.
| Benazepril-Hypertension as ?-SARS |
|---|
| peramivir |
| tamiflu17 zanamivir9 |
| gilteritinib fumarate |
| rimantadine hydrochloride11,12 |
| benazepril hydrochloride doravirine galantamine hydrobromide cetirizine hydrochloride hives lanadelumab |
| aliskiren hemifumarate30 |
| desloratadine entacapone invirase daclatasvir dihydrochloride indacaterol maleate loratadine peganone |
| nitazoxanide31 |
| denavir triprolidine and pseudoephedrine hydrochlorides codeine rivastigmine telavancin hydrochloride donepezil hydrochloride triprolidine and pseudoephedrine hydrochlorides tazemetostat hydrobromide |
| relenza9 |
| benazepril hydrochloride and hydrochlorothiazide nulojix ecallantide alectinib hydrochloride |
| virazole29 |
| levocetirizine hydrochloride |
| donepezil hydrochloride and memantine hydrochloride11,12 amantadine hydrochloride11,12 |
| cetirizine hydrochloride comtan |
| fluvoxamine maleate32 amlodipine besylate and benazepril hydrochloride33 |
| delafloxacin meglumine acrivastine dalbavancin hydrochloride |
| fexofenadine hydrochloride hives26 |
| rilpivirine hydrochloride aricept bendamustine hydrochloride viramune xr revefenacin olodaterol hydrochloride meloxicam |
Hits containing drugs suggested or under investigation for COVID-19 are highlighted in gray.
| Albuterol-Asthma as ?-SARS |
|---|
| peramivir albuterol albuterol sulfate albuterol sulfate and ipratropium bromide |
| zanamivir9 rimantadine hydrochloride11,12 |
| pralidoxime chloride meperidine and atropine sulfate |
| amantadine hydrochloride11,12 |
| doxacurium chloride biperiden lactate atropine sulfate syringe gallamine triethiodide atropine and demerol colistin sulfate |
| oseltamivir phosphate17 |
| revefenacin dextromethorphan hydrobromide and quinidine sulfate conivaptan hydrochloride glycopyrronium tosylate cefiderocol sulfate tosylate fentanyl citrate and droperidol pancuronium bromide |
| relenza9 |
| telavancin hydrochloride guaifenesin and dextromethorphan hydrobromide diphenoxylate hydrochloride and atropine sulfate |
| esketamine hydrochloride34 |
| galantamine hydrobromide naloxone hydrochloride and pentazocine hydrochloride |
| glycopyrrolate35 |
| levalbuterol hydrochloride calfactant rilpivirine hydrochloride pipecuronium bromide |
| tamiflu17 |
| biperiden hydrochloride mivacurium chloride metocurine iodide ceftolozane sulfate atropine sulfate terbutaline sulfate nesiritide recombinant diphenoxylate hydrochloride atropine sulfate tubocurarine chloride benzonatate rapacuronium bromide naloxone hydrochloride propoxyphene hydrochloride and acetaminophen acetaminophen and pentazocine hydrochloride |
Here we present results for validation of our word embedding mining approach along with results from applying our approach for COVID-19 drug repurposing. First, we present validation results for our approach to ranking FDA approved drugs for three diseases or disease families with well-established treatments. Specifically, we use the same three seed drug-disease pairs as analogies to find drugs for Alzheimer’s, allergies, and cancer (see Table 1, Table 2, and Table 3). All drugs with a primary indication for the query disease are highlighted in gray. This is to verify that our complete approach (drug vectors ranked by cosine similarity to treatment analogy vector) can identify effective ground-truth drugs for diseases that are not closely related to the seed disease-drug pair. In nearly every example, a vast majority (if not all) of the top 10 hits have a primary indication for the query disease.
Next, we present the 50 closest FDA approved drugs to the treatment analogy vectors for SARS, thereby filtering to what may be the most promising drugs for repurposing. The top repurposing hits are presented in Table 4, Table 5, and Table 6, and all drugs that have been suggested for or are currently under investigation for treatment of COVID-19 are highlighted in gray. This highlighting serves as a partial evaluation of the repurposing via positive controls, suggesting that other hits may be good candidates for further investigation. We find 22 positive control hits out of 50 for the metformin-diabetes analogy, 12 of 50 for the benazepril-hypertension analogy, and eight of 50 for the albuterol-asthma analogy. We present a Venn diagram of the overlap between the three analogies in Figure 1, and a table containing the drugs shared by all three and by at least two of the analogies in Table 7. Seven drugs are shared by all three analogies in their top 50 hits, and another 10 are shared by at least two of the analogies for a total of 17 higher confidence hits.
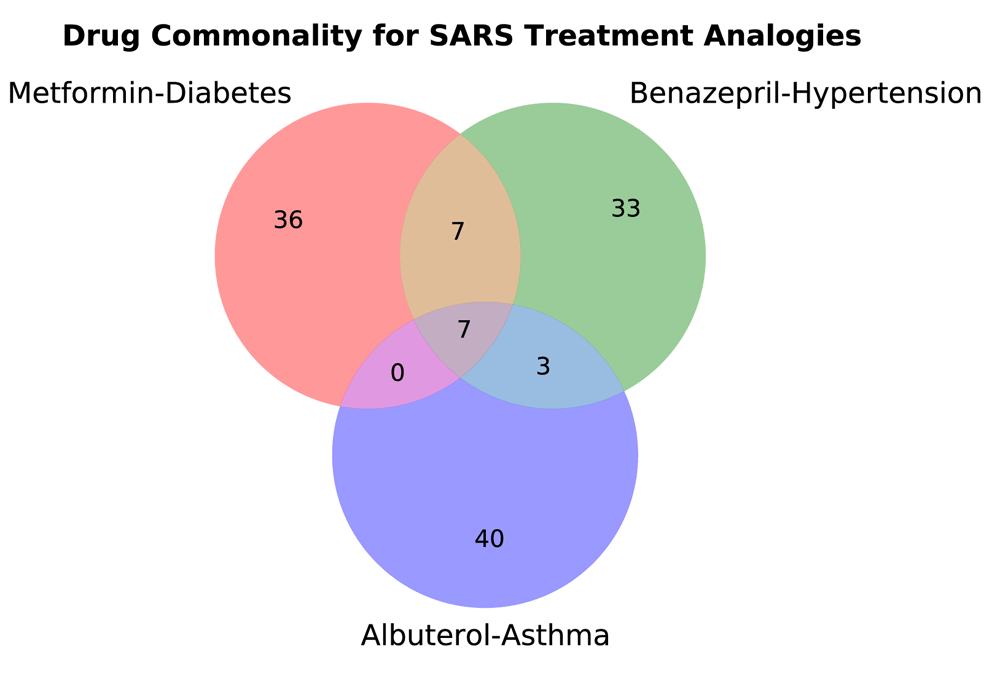
| Drug Repurposing Candidate Commonality for SARS | |
|---|---|
| Common to all | amantadine hydrochloride11,12 |
| peramivir revefenacin rilpivirine hydrochloride | |
| rimantadine hydrochloride11,12 tamiflu17 zanamivir9 | |
| Common to two | alectinib hydrochloride |
| donepezil hydrochloride and memantine hydrochloride11,12 | |
| doravirine galantamine hydrobromide gilteritinib fumarate | |
| relenza9 | |
| telavancin hydrochloride triprolidine and pseudoephedrine hydrochlorides triprolidine and pseudoephedrine hydrochlorides codeine | |
| virazole29 | |
Here we review the validation results to demonstrate that our approach can find useful drugs for various diseases, followed by manual review of the FDA approved drug repurposing candidates for SARS. First, recall that we have used our drug ranking approach with the same seed analogy vectors for three major diseases with well-established ground-truth treatments. For the validation of our approach on drugs for Alzheimer’s, nearly all of the drugs suggested from each analogy were drugs with primary indications for Alzheimer’s, and several of the seemingly incorrect drugs have a primary indication for Parkinson’s, another neurodegenerative disease. We see a similar result for allergies where only the albuterol-asthma analogy suggests drugs not indicated for allergies in the top 10. Specifically, we see albuterol and levalbuterol show up several times, perhaps as a result of seed drug bias. For the cancer drugs, we see that every drug is indicated for some form of cancer. All of this reassures us that our approach does, in fact, find drugs appropriate for the query disease even if the query disease has no relationship with the seed drug-disease pair.
Next, we manually reviewed every one of our top 50 FDA approved drugs suggested for repurposing with SARS as the query disease, and marked every one that has either been suggested for or is currently under investigation for treatment of SARS-CoV-2 and COVID-19. From the metformin-diabetes analogy, we find 22 of 50 drugs either suggested or under investigation for treatment against SARS-CoV-2 and COVID-19. With the benazepril-hypertension analogy, we find 12 of 50 hits, and from the albuterol-asthma analogy, we find eight of 50. Across the analogies, seven hits are common to all three, and 10 are common to two of the three.
In the seven hits common to all, four have been suggested for treatment of SARS-CoV-2 and COVID-19. Amantadine and rimantadine are both adamantanes, which have been shown to have antiviral properties in vitro and have demonstrated possible protective effects in a clinical study of patients with neurological diseases11,12. Zanamavir is an antiviral that has been suggested based on in silico molecular docking models of the 3C-like proteinase9, which is a major protease thought essential to viral replication of coronaviruses, including SARS-CoV and SARS-CoV-236,37. Oseltamivir (Tamiflu) is another antiviral that is under investigation via clinical trial17.
In the 10 hits common to two of the analogies, three have been suggested for treatment of SARS-CoV-2 and COVID-19. Memantine is another adamantane similar to amantadine and rimantadine suggested by all three analogies. Relenza is a trade name for zanamivir, so is essentially a duplicate, though it does perhaps suggest even more confidence in the drug. Virazole is a trade name for ribavirin, an antiviral which has shown antiviral activity against SARS-CoV-2 in vitro29.
We also note that 13 of all the proposed treatments are in clinical trials: atovaquone, lopinavir and ritonavir, sirolimus (suggested here as the prodrug temsirolimus), oseltamivir, selinexor, ibuprofen, colchicine, bicalutamide, mefloquine, chloroquine, linagliptin, fluvoxamine, and ketamine (suggested here as the enantiomer esketamine). Interestingly, these drugs come from a wide range of primary indications including antiparasitic, antiviral, anti-inflammatory, anticancer, anesthetic, and antidepressant effects. Furthermore, the proposed drugs that are not currently in trials show a similar breadth of primary indication. Overall, we find that our approach shows a great deal of promise as it is able to discover a wide range of drugs that have elsewhere been proposed for COVID-19 from clinical, in silico, in vitro, and in vivo experimentation, all done here with literature published before SARS-CoV-2 was discovered.
Of course, while our method appears promising, it is not without limitations. First, our method is limited to what has already been published in the scientific literature and cannot propose new drugs or treatments outside of the embedding vocabulary. We also caution readers that, in most cases, these drugs have not been tested for COVID-19 efficacy, and we make no claims other than that some of these drugs deserve further exploration. We can say with confidence that at least a few proposed drugs seem less promising. Peramivir is a neuraminidase inhibitor used to treat influenza. While it is thus an antiviral, coronaviruses do not use neuraminidase, so it would seem less likely to be effective against SARS-CoV-222. On the other hand, zanamivir and oseltamivir, two of our common positive controls9,17, are also neuraminidase inhibitors and should thus be less likely candidates. Given that the potential mechanism of action for zanamivir at least is based on computed binding to the 3C-like proteinase, perhaps some drugs may demonstrate efficacy outside of their traditional mechanism. Nevertheless, the lesson is that we should expect to find false positives in our top hits along with any true positives. Finally, our embedding approach does not take into account the potential of drug-drug interactions to increase or decrease efficacy in any fashion. All of this is to say that further in vitro and in vivo experimentation, and observational EHR or claims data would all be useful additional sources of evidence for or against repurposing candidates listed here.
In this work, we present a word embedding mining approach to identifying candidate treatments for SARS-CoV-2 and COVID-19. We first use seed drug-disease pairs to produce treatment analogy vectors for a query disease using a prebuilt biomedical word embedding. We then use a simple word vector averaging approach to get vectors for a list of FDA approved drugs and sort them by their distance to our treatment analogy vectors. We validate that this approach identifies ground truth treatments for well-known diseases. Next, we use the same approach to produce a list of candidate drugs for the query disease SARS, manually evaluate the top candidate drugs, and find several positive controls that have been suggested in the literature or are currently under investigation for SARS-CoV-2 or COVID-19 treatment. While there are certain to be several false positives amongst our top hits as well, we find the presence of positive controls reassuring, and propose the remainder as potential candidates for further investigation. We furthermore propose this word vector embedding approach in general as a useful tool for COVID-19 drug repurposing. These results only scratch the surface of what is possible and we present this work as a suggestion to the community to investigate further. Immediate avenues for future investigation include exploring even more drug-disease analogy vectors, ranking drugs directly by their cosine similarity to proven treatments as they arise, and investigating drug-gene target analogy vectors rather than the disease treatment analogy we demonstrate here.
The FDA database of approved drugs is available at: https://www.fda.gov/drugs/drug-approvals-and-databases/drugsfda-data-files.
All code and processed data used to produce these results are available at: https://github.com/finnkuusisto/covid19_word_embedding.
Archived code and data as at time of publication: http://doi.org/10.5281/zenodo.38600574.
License: CC0
The code is provided in Python (v 3.8) as Jupyter Notebooks (v 6.0.3), and additionally requires Gensim (v 3.8.1), Matplotlib (v 3.2.1), and Matplotlib-Venn (v 0.11.5).
The BioWordVec prebuilt embedding is available via the official GitHub repository: https://github.com/ncbi-nlp/BioWordVec.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the work clearly and accurately presented and does it cite the current literature?
Yes
Is the study design appropriate and is the work technically sound?
Partly
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
Not applicable
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Deep Learning, computational drug repurposing, translational bioinformatics, EHR
Is the work clearly and accurately presented and does it cite the current literature?
Partly
Is the study design appropriate and is the work technically sound?
Yes
Are sufficient details of methods and analysis provided to allow replication by others?
Partly
If applicable, is the statistical analysis and its interpretation appropriate?
I cannot comment. A qualified statistician is required.
Are all the source data underlying the results available to ensure full reproducibility?
Yes
Are the conclusions drawn adequately supported by the results?
Partly
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Computational biology, system biology
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 10 Jun 20 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)