Keywords
Coronavirus, COVID-19, SARS-CoV-2, knowledge graph, knowledge base, target discovery, knowledge mining, bioinformatics
This article is included in the Bioinformatics gateway.
This article is included in the ELIXIR gateway.
This article is included in the Emerging Diseases and Outbreaks gateway.
This article is included in the Coronavirus (COVID-19) collection.
Coronavirus, COVID-19, SARS-CoV-2, knowledge graph, knowledge base, target discovery, knowledge mining, bioinformatics
The global COVID-19 pandemic, caused by the SARS-CoV-2 virus, has infected millions worldwide, causing many more infections and deaths than the previous severe acute respiratory syndrome (SARS) outbreak that occurred in 2002 and 2003 (WHO, 2020). The race to find plausible treatment strategies and management plans for the coronavirus pandemic, concurrent with investigating the biological underpinnings of how SARS-CoV-2 operates has driven a very rapid rise in the number of SARS-CoV-2-related publication, pre-prints and biological data. The White House, alongside other groups, made a global call to action to find a way to rapidly sift through this mountain of COVID-19 literature. They released the COVID-19 Open Research Dataset (CORD-19), which has over 150,000 COVID-19-related full text articles (Kaggle, 2020). Additionally, the COVID-19 data portal contains over 360,000 publications and many other datasets related to the disease. The rate of growth and sheer volume of data related to the pandemic and the virus has become so large that it has led to something of a new phenomenon for many researchers: ‘too much information’. As such, there lies a challenge to organize the growing mountain of biomedical knowledge in order to overcome the data interpretation bottleneck (Good et al., 2014).
Our goal was to repurpose the KnetMiner software, which we originally developed for crop scientists to help identify the most important genes involved in complex plant traits (Hassani‐Pak et al., 2021), to provide medical researchers with quick and intuitive access to all documented linkages between genes, potential therapeutic compounds, and the virus. KnetMiner contains fast algorithms for scanning millions of relationships from across a range of datasets and literature, scoring all evidence and displaying the links in easy to understand knowledge graphs. KnetMiner for COVID-19 would enable users to search for genes and keywords related to COVID-19 and explore the surrounding connected gene networks and pathways, for example, for negative downstream effects of drugs, or genetic associations with known diseases, or human-pathogen interactions.
KnetMiner requires as input a knowledge graph (KG) which is a flexible semantic data model to represent heterogeneous, interconnected data where graph nodes represent biological concepts and graph edges the relationships between them. Such biological concepts can include, but are not limited to, genes, drugs, diseases, and publications. There have been a few parallel efforts to construct KGs from COVID-19 data (Reese et al., 2020). These mostly differ in data coverage and modelling approaches and would require further work to be made usable by KnetMiner. Hence, we decided to use the toolkit provided by KnetMiner to build a unified knowledge graph from public datasets and the scientific literature.
Here, we describe how we organized the biomedical knowledge in a format that is compatible with KnetMiner and present a use case.
Firstly, we identified the datasets that we considered as important for the first iteration of the COVID-19 KG, based on discussions held with participants of the COVID-19 Biohackathon (5–11 April 2020). We downloaded over 20 datasets including COVID-19 related publications and pre-prints, human genetics studies, virus-host protein interactions, drug-target interactions, pathway information, ontology annotations and mouse knock-out mutant data with links to diseases and phenotypes (see Table 1). The data download and processing steps were automated using custom Python scripts (see Extended data). Various data formats are supported by the down-stream integration pipeline, including PubMed XML, UniProt XML and OWL, and most importantly tabular format, which required the development of an XML-based configuration to map the tabular content to a labelled property graph representation.
The KnetBuilder (https://github.com/Rothamsted/knetbuilder) software was used to integrate the various data sources into a KG with unified semantics and augment it with additional text mining-based relationships (Hassani-Pak et al., 2010). The data loading and integration steps are specified using an XML-based configuration. KnetBuilder uses an in-memory graph implementation (Köhler et al., 2006) and provides exports to RDF and tooling to convert RDF to Neo4j (Brandizi et al., 2018b).
KnetMiner is an open-source software allowing to index and serve the contents of a knowledge graph in order to accelerate gene discovery. It requires as input a dataset folder with configuration files and a compatible knowledge graph in OXL format. We used the version 4.0 of KnetMiner and deployed it with our COVID-19 KG (see Underlying data). The COVID-19 KG contains all human genes; however, KnetMiner was configured to index a subset of 9,141 COVID-19-related genes, derived from SciBite gene annotations of CORD-19 version 54 (Giles et al., 2020). In the presented use case, KnetMiner was searched with a list of differentially expressed genes (Table 3) and the following keywords:
"Defense Response To Virus" Remdesivir "Killing Of Cells Of Other Organism" "Antimicrobial Humoral Immune Response Mediated By Antimicrobial Peptide" "Response To Virus" "Cellular Response To Lipopolysaccharide" "Positive Regulation Of Transcription By RNA Polymerase II" "White blood cell count" "Blood protein levels"
ConceptID:500458 ConceptID:518058 ConceptID:511770 ConceptID:513484 ConceptID:503458 ConceptID:512650 ConceptID:471625 ConceptID:379514 ConceptID:370288
The combined COVID-19 KG has 674,969 biological concepts and over 1.6 million relationships between them, and is available in OXL, RDF and Neo4j formats. It contains data on 27,599 human genes that are present in the KG as “type gene” concepts that are linked to a range of other concepts through different relation types and evidences. The content of the KG with a breakdown by concept type is shown in Table 2.
The most up-to-date statistics can be accessed at: https://knetminer.com/COVID-19/html/release.html
Programmatic access is available via public SPARQL and Cypher endpoints (Brandizi et al., 2018a) (see Underlying data). We have developed sample Jupyter notebooks to illustrate how the KG can be queried (Brandizi, 2020). This maximizes the adhesion to FAIR data principles, ensuring the data is reusable for a variety of applications. Availability, including pointers to data repositories on the Open Science Framework (OSF), is listed in the README file (see data availability section). Using Neo4j and Cypher, we can query the data and walk the knowledge graph in multiple directions. For example, we can look at the most common pathways related to proteins which are targeted by literature cited drugs using the following Cypher query:
COVID-19 KnetMiner (https://knetminer.com/COVID-19/) provides gene-centric and interactive access to the integrated knowledge base created by KnetBuilder. KnetMiner can be used to search the KG with keywords, gene lists, genomic regions of interest, and/or any combinations of these user inputs. We have created several example queries for different use cases and search strategies. These are available on the COVID-19 KnetMiner website for easy use.
In June 2020, a gene expression study was published which investigated the transcriptional response of human lung epithelial cells to SARS-CoV-2 infection (Enes & Pir, 2020). The data and the analysis between infected vs mock cells are available at the EBI Gene Expression Atlas (Table 3; https://www.ebi.ac.uk/gxa/experiments/E-GEOD-147507).
We chose this dataset because of its relevance and tractable size to showcase the use of KnetMiner for the interpretation of differentially expressed genes (DEGs). The approach can be equally applied to any data driven analysis and derived gene lists. All DEGs from the above study were used in KnetMiner to investigate linkages to drugs, diseases, biological processes, and the CORD-19 literature. As such the DEG IDs were entered into the Gene List search interface of KnetMiner and analyzed in several iterations (Figure 1): (i) without any keywords to get an unbiased view of the linked and enriched knowledge, (ii) with the keyword SARS-CoV-2 to review recent CORD-19 articles in relation to these genes, and (iii) with specific drugs, diseases and GO terms to visualize and share the organized knowledge with other scientists (Figure 2).
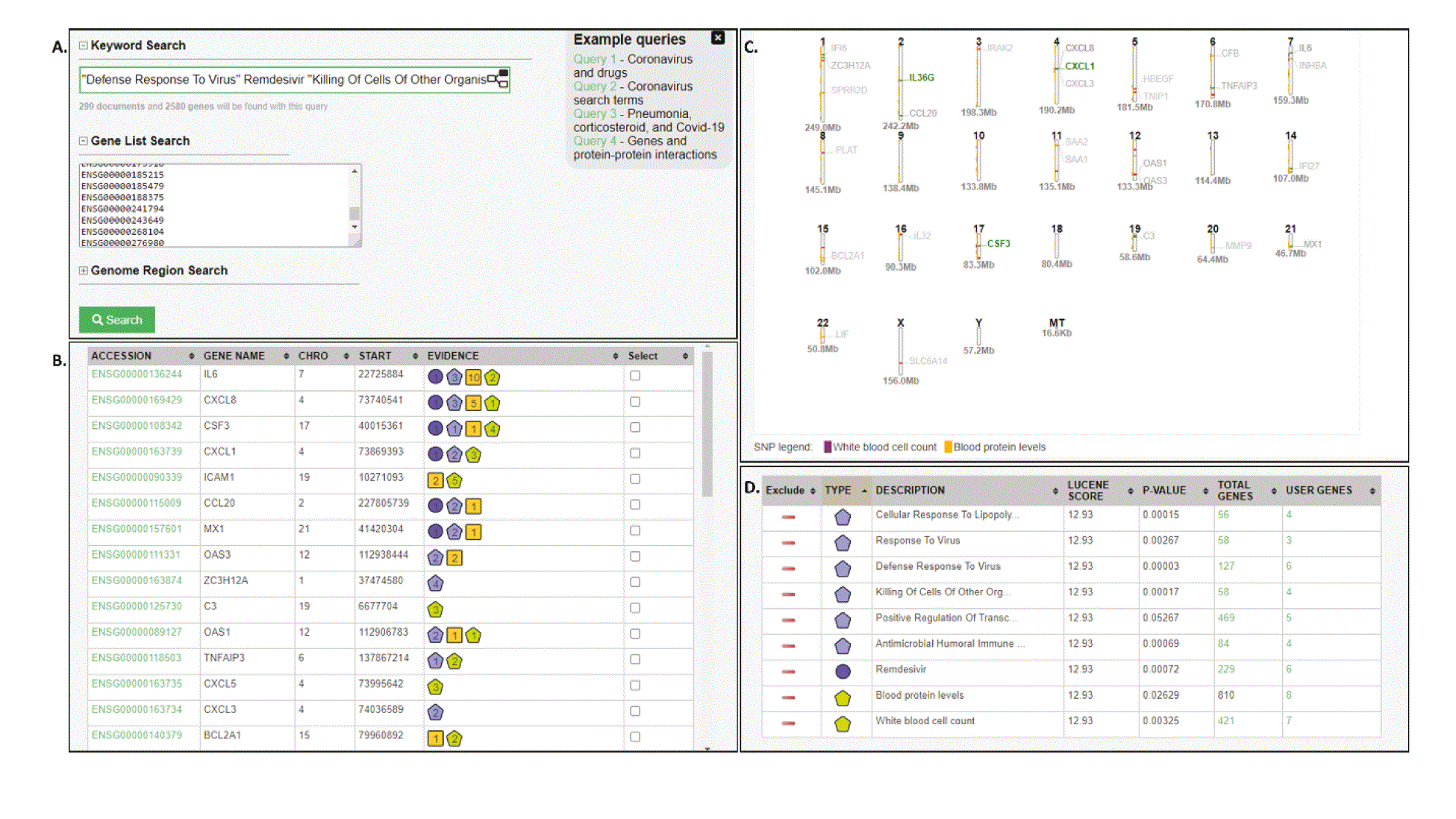
A) Search result for gene list and search terms. B) Gene View: Ranked list of genes and evidence summaries. C) Map View: Interactive genomic map with related GWAS studies and top scoring genes. D) Evidence View: Enriched linked knowledge.
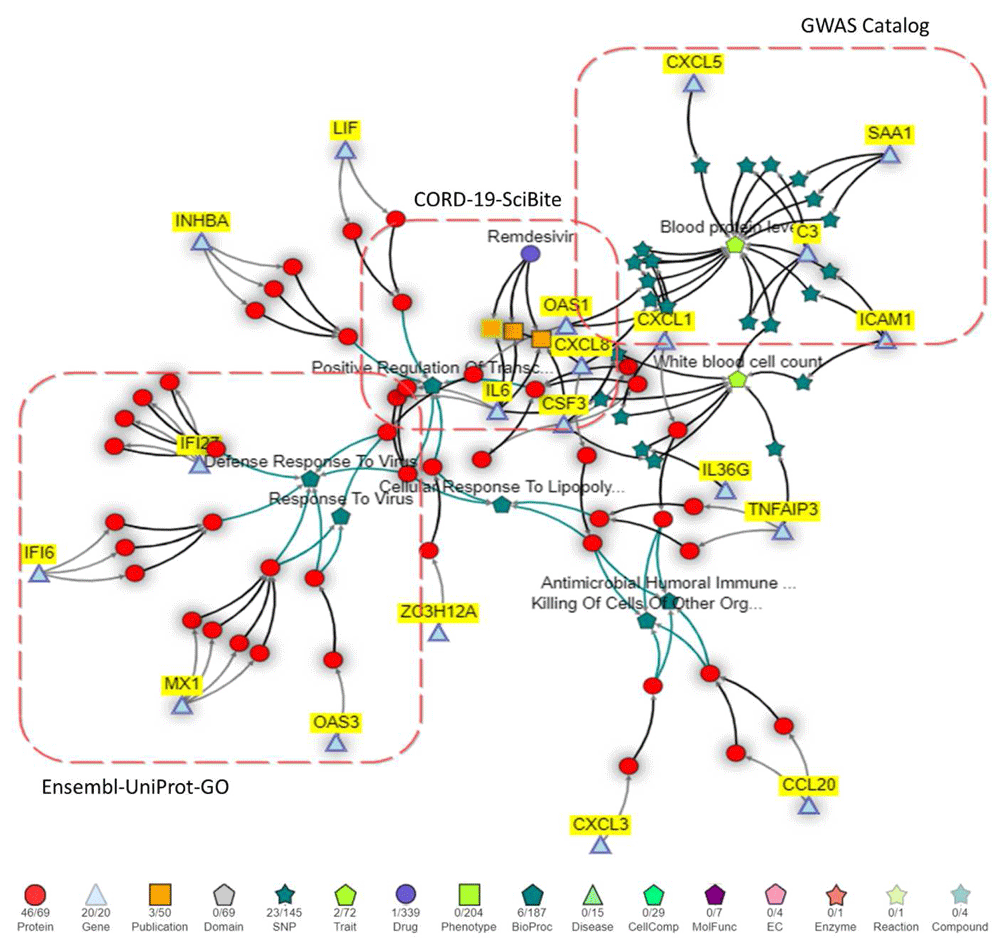
Information from various sources are visualized in a single view. Users can add or hide information using the legend. In total, the network contains 1216 nodes and 1808 edges, of which 101 and 143 are shown respectively. For example, 2 out of 72 Trait concepts are visible and other linked Traits (GWAS studies) can simply be added by double-clicking the Trait symbol in the legend.
KnetMiner user stories usually follow a common pattern: search, prioritize, explore, and share knowledge. All views shown in Figure 1 can generate knowledge networks for selected genes and/or evidence terms. As an example, an interactive network was generated for the top 20 scoring DEGs and keywords related to white blood cell count (GWAS study), Remdesivir (Drug), Response to Virus (GO).
KnetMiner provides open-source tools to build, search, visualize and share knowledge graphs, and its species agnostic architecture has provided a cost-efficient toolkit for building the first COVID-19 KnetMiner resource. By organizing COVID-19 related data in one place, integrated through a clear semantic data model, the hope is that this will enable data interpretation using more reproducible and objective approaches and support the international search for useful drugs, stop researchers repeating work done elsewhere, avoid harmful interventions, and ultimately, help pave the way to effective treatments. In an effort to maximize our data usefulness and grow the knowledge graph faster, we plan to offer mappings and conversions between our data and the KGX/Biolink ecosystem (Biolink, 2020). For instance, we are investigating methods to ingest the covid-19-kg produced by the Berkely Lab (Reese et al., 2020) and integrate it into the KnetMiner platform. This is work that will be conducted within the COVID-19 international research team (COV-IRT).
COVID-19 KG: https://github.com/Rothamsted/covid19-kg/
RDF Endpoint: http://knetminer-data.cyverseuk.org/lodestar/sparql
Neo4j Endpoint: http://knetminer-covid19.cyverseuk.org:7476/browser/
Documentation and custom scripts are available from https://github.com/Rothamsted/covid19-kg/ (see README)
Archived scripts as at time of publication: https://doi.org/10.5281/zenodo.5094695
License: AGPL-3.0
KnetMiner web app: https://knetminer.com/COVID-19
ELIXIR bio.tools: https://bio.tools/covid-19_knetminer
Software available from: https://hub.docker.com/u/knetminer
Source code available from: https://github.com/Rothamsted/knetminer
Archived source code at time of publication (usually Zenodo): https://doi.org/10.5281/zenodo.3891097
License: AGPL-3.0
We are grateful to the organizers of COVID-19 Biohackathon and the participants of the Knowledge Graph topic, especially Justin Reese, Deepak Unni and the SciBite lab for their support with our queries. We would also like to thank the COVID-19 International Research Team (https://www.cov-irt.org/) for allowing us to present KnetMiner and for providing an excellent networking channel. We additionally thank CyVerse UK and acknowledge UKRI BBSRC support (BB/R000662/1) for providing us with resources to host our Neo4j and RDF COVID-19 knowledge graphs.
| Views | Downloads | |
|---|---|---|
| F1000Research | - | - |
|
PubMed Central
Data from PMC are received and updated monthly.
|
- | - |
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Natural Language Processing, Machine Learning
Is the rationale for creating the dataset(s) clearly described?
Yes
Are the protocols appropriate and is the work technically sound?
Yes
Are sufficient details of methods and materials provided to allow replication by others?
Partly
Are the datasets clearly presented in a useable and accessible format?
Yes
Competing Interests: No competing interests were disclosed.
Reviewer Expertise: Knowledge graphs, machine learning, data science
Alongside their report, reviewers assign a status to the article:
| Invited Reviewers | ||
|---|---|---|
| 1 | 2 | |
|
Version 1 30 Jul 21 |
read | read |
Provide sufficient details of any financial or non-financial competing interests to enable users to assess whether your comments might lead a reasonable person to question your impartiality. Consider the following examples, but note that this is not an exhaustive list:
Sign up for content alerts and receive a weekly or monthly email with all newly published articles
Already registered? Sign in
The email address should be the one you originally registered with F1000.
You registered with F1000 via Google, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Google account password, please click here.
You registered with F1000 via Facebook, so we cannot reset your password.
To sign in, please click here.
If you still need help with your Facebook account password, please click here.
If your email address is registered with us, we will email you instructions to reset your password.
If you think you should have received this email but it has not arrived, please check your spam filters and/or contact for further assistance.
Comments on this article Comments (0)